Whether you’re designing novel antibodies for your groundbreaking vaccine work or you’re breaking down a key pathway in cancer metastasis, you know the importance of understanding how the components of your system are interacting. At this point you have achieved great results from the preliminary stages of your interaction research workflow – you’ve identified a key protein in your system, and it has been successfully expressed and purified. You are now ready to begin investigating how your protein interacts with the other components of your system, starting by identifying binding partners of interest.
At this stage, techniques such as Co-immunoprecipitation (Co-IP) and native electrophoresis are typically employed to isolate interacting partners so that they can be identified using mass spectroscopy. These techniques are an efficient and cost-effective way to qualitatively confirm binding between your target protein and its interacting partners. However, more and more reviewers are demanding stronger data than just yes/no binding. Reporting quantitative binding kinetics data gives the whole picture behind the interaction taking place, which significantly increases your credibility when it’s time to publish.
So, what tools do you have available to achieve this? While several techniques exist, such as ITC, MST, and BLI, surface plasmon resonance (SPR) is one of the most versatile approaches due to the wide variety of data that can be collected about your interactants using a single instrument. Below we present four different types of binding assays that can all be conducted using SPR.
1. Binding Kinetics Assay
If you have been keeping up with our other blog posts, you already know why surface plasmon resonance is a powerful method for measuring the on rate, off rate, and overall affinity of your interaction. Keep in mind that an effective binding kinetics assay requires careful preparation and optimization to produce publication-quality data. For our purposes, we will quickly summarize what a typical experiment looks like.
Assuming that you have two purified binding partners ready to go, it is now time to choose one your of interactants to be immobilized to a sensor chip of appropriate surface chemistry – this will be your ligand. Your other interactant (the analyte) is then injected and flows over your immobilized ligand in a buffer at 3-5 different concentrations. At this point, you can use SPR data analysis software to fit your data to an appropriate model and calculate the KOn, KOff, and KD of your interaction.

Figure 1. Visualization of the ligand (binding partner immobilized to the sensor surface) and the analyte (binding partner in solution).
To reiterate, there are many other factors to consider when designing a binding experiment, meaning optimization is not always trivial. Fortunately, anybody using an OpenSPR instrument has access to our amazing Customer Support Scientists who are there every step of the way to get your experiments optimized and help you produce beautiful kinetics data.
2. Competition Assay
Now that we have seen a straightforward binding kinetics assay, we can now explore a similar assay that addresses a unique challenge in the field of drug discovery. Measuring a binding interaction between two compounds with a large size difference is challenging with any technique, and when working with small molecule drugs, this is a common problem. Fortunately, the KD of a protein-small molecule interaction can be measured indirectly using a competition assay.
This technique requires a different binding partner of your target protein that does not interact with your small molecule. This alternative ligand is immobilized, and a standard binding kinetics assay is performed using your target protein as the analyte.
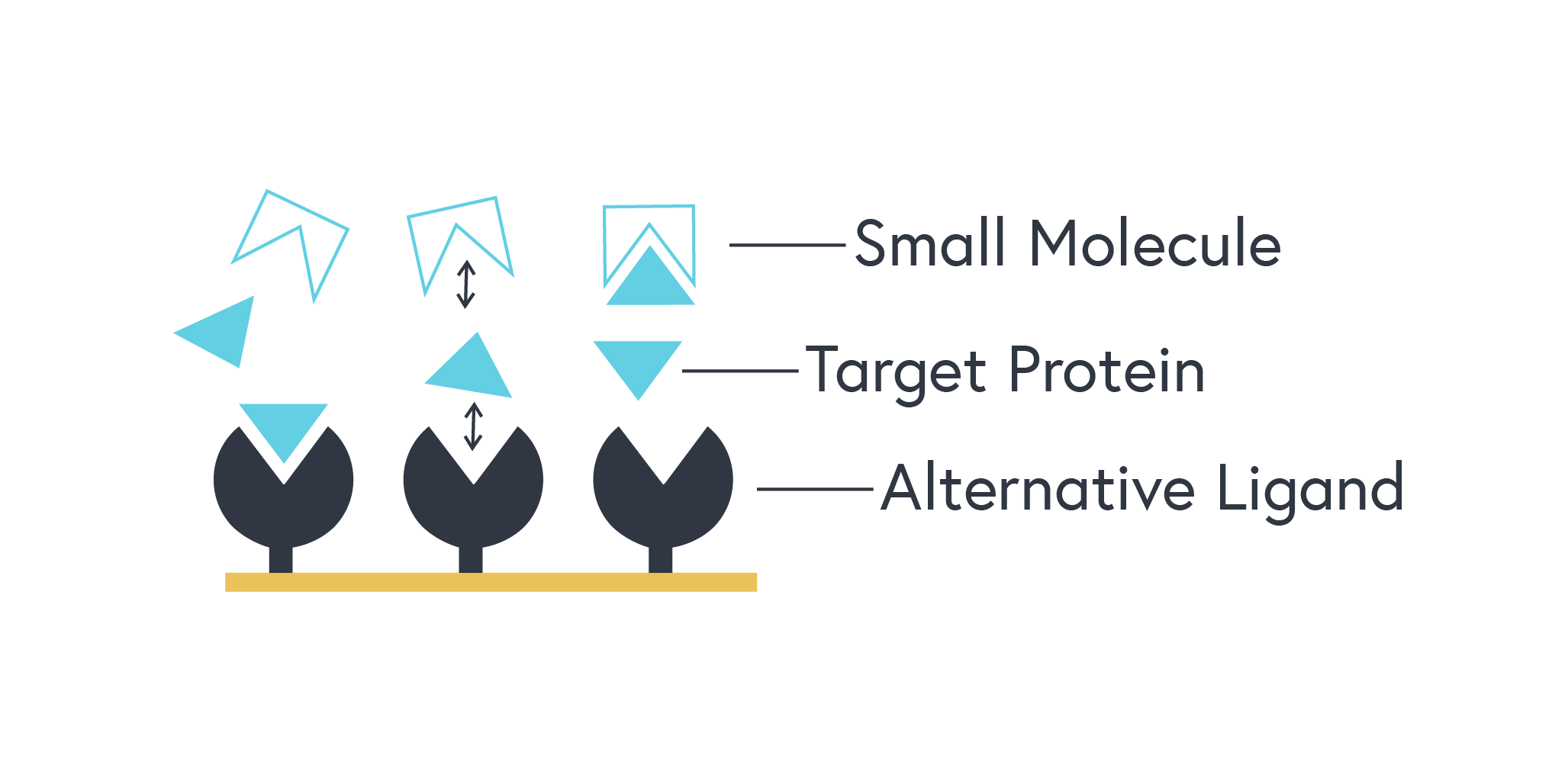
Figure 2. Representation of a competitive assay: The target protein, initially free to bind the alternative ligand, will bind the small molecule instead as it is introduced. This will be observed by a reduction in signal from the instrument.
From the data collected, you will be able to use post-processing software to calculate a relative KD for the protein-small molecule interaction. It is important to note that a competition assay does not provide KOn and KOff, only the overall affinity of the interaction. That being said, with this technique, you can now indirectly calculate the KD of a protein-small molecule interaction regardless of the size difference of your molecules.
3. Thermodynamics Assay
So far, we have been keeping the temperature constant, but we know that kinetics is dependent on temperature. Using a thermodynamics assay we can determine the effects of temperature on our interaction and quantify several other biophysical properties as well. Remember that there are potential complications when playing with heat, such as protein denaturation at higher temperatures.
The assay here is straightforward. You set up a binding kinetics assay as described above, only this time, your analyte concentration is kept constant, and the temperature varies between steps. Without any additional math, you can induce a lot about the effects of temperature on your interaction based on the change in KD you observe with temperature. If we want to calculate more, we now must do some physical chemistry calculations. Without going into too much detail, here is a breakdown of the calculation:
KD is subbed in for Ka in the equation: ΔG = -RTln(Ka)
This equation is then subbed into: ΔG = ΔH – TΔS
The resulting equation is rearranged into the form: ln(KD) = ΔH/(RT) – ΔS/R
This is now in the linear form: y = mx + b, where y = ln(KD) and x = 1/T
By plotting this using our experimental data, the resulting linear equation gives us: m = ΔH/R and b = -ΔS/R.
Through some simple rearranging, we can now solve for the change in enthalpy, entropy, and Gibb’s free energy resulting from our interaction. This powerful assay allows us to obtain even more biophysical data for our interaction using the experimental conditions we already optimized when obtaining binding kinetics.
4. Concentration Assay
With one optimized experiment, we can now calculate full binding kinetics (KOn, KOff, KD) as well as ΔG, ΔS, and ΔH using tools from our interaction toolbox. So, what else can we do with this experiment? Another interesting application with SPR is a concentration assay. This type of assay determines where the concentration of your protein of interest is in an unknown sample. While this is an effective method for analyzing crude samples, it is important to note that if any other components of the unknown sample bind specifically with your ligand or non-specifically with your sensor chip, the results can be skewed.
First, you perform a traditional binding assay for interaction with your target protein that is well documented with high reproducibility to ensure accuracy. Using your target protein as the analyte, the maximum signal at each concentration is plotted against your target protein concentration to obtain a standard curve. One more experiment is then run using the same ligand immobilized, but now the unknown sample is the analyte. The maximum signal from this experiment is plugged into the linear equation of your standard curve to solve for the concentration of your target protein.
Conclusion
As quantitative biophysical data becomes a standard in publications, it’s crucial to know what tools you have available to provide the data you need. With surface plasmon resonance you can measure a variety of biophysical data in real-time, without the use of labels, and without excessive sample consumption. With the OpenSPR this technique has never been more affordable and accessible to researchers, and our Customer Success Scientists are there every step of the way to help.
So what binding assay is the best fit for your research? Our Application Scientists are always available to discuss your applications, determine what approach is best for you, and help you achieve the ultimate goal of this entire process – getting published faster.
