Trends in Publications
How do you measure success as an academic researcher? Although there are many paths that a scientific research project takes, the end goal remains the same; publishing your work. Our mission at Nicoya is to help researchers publish faster with high-quality quantitative binding data. As the availability of affordable technology increases, reviewers are constantly requesting quantitative binding data. Although there are many available methods and technologies that could provide you with quantitative binding data, they may not satisfy all requirements you need to successfully publish your paper. In this article, we describe a general outline of a research workflow and how you can incorporate quantitative binding data to successfully publish your work. This outline may not completely resemble your project milestones and timelines, as each research project is unique. To learn more about incorporating quantitative binding data to your specific research project, speak to an application scientist here.
Preliminary Research
Starting any major research project begins with the identification of a problem to study (ie. cancer). This involves countless hours of reading existing research and locating gaps in the current literature (for ex. identifying an under researched protein involved in a known metastatic pathway). This process can take many months and will likely result in the submission of a grant application to institutions such as the NIH or NSF. Obtaining preliminary data using in silico methods (ie. SwissPDB Viewer, UniProt) may be employed during this stage to begin reviewing protein structure and/or finding potential interactors.
Protein Expression and Purification
At this point, your protein of interest has been identified, your grant has been funded and it’s time to begin the labour, with the help of your talented lab members. Simply put, in this project stage, your protein of interest is “manufactured” using cellular machinery to further carry out all of the necessary downstream assays. A stock culture of an expression system (ie. E. coli) must be cultivated so you can subsequently recombine the gene encoding your protein of interest into a plasmid and transform your cells to overexpress this protein of interest.
Following the aforementioned methods, you may begin to sonicate, differentially centrifuge and concentrate your protein of interest from the overexpressing cells you’ve produced during upstream processing. Methods such as column chromatography (ie. Size Exclusion Chromatography, Ion Exchange Chromatography and/or Affinity Chromatography) may be employed sequentially to purify your protein of interest for downstream assays.
Gel Electrophoresis and Western Blotting
As scientists, we all know how important it is to have appropriate quality control measures in place and in this respect, validation that your protein of interest is successfully purified must be accomplished. Gel electrophoresis will separate your protein of interest based on the size and charge and provided that the size of your protein is known (ie. from UniProt), you will be able to confirm the success of your prior experiments. Moreover, western blots can be conducted to further confirm the presence of your protein of interest with the employment of protein-specific antibodies; note that western blots should be carried out at various points in your workflows and not just in this project stage! Upon successful western blots, it’s time to identify potential protein binding partners.
Identifying Protein Binding Partners
Upon expression, purification and confirmation of the presence of your protein of interest, it’s time to find potential interacting protein partners. Co-immunoprecipitations (or pull-down assays) are the most common way to find interacting partners with your protein of interest. These techniques will provide yes/no, qualitative binding data but will not quantitate the strength and rate of the interaction; this is where the importance of quantitative information begins to become important.
Characterizing Protein Binding Partners
The techniques we’ve described above are very standard and traditional methods, but in order to stay ahead of reviewers, it’s vital to incorporate more comprehensive and quantitative technologies into your projects. With that said, there are a few techniques that can provide quantitative binding data including ITC, MST and BLI. However, these technologies typically use a lot of your sample, require fluorescent labels and/or large capital expenditure. Surface Plasmon Resonance (SPR) is known to be the gold standard for obtaining quantitative binding kinetics data and at this point in your research project, your protein of interest and its respective binding partners can be characterized with binding kinetics. Employing SPR in your research workflow will help you keep reviewers happy and publish your work faster.
Why is SPR critical for publications? How does OpenSPR help?
SPR is a label-free technology which allows researchers to quantitatively analyze binding between two biomolecules. SPR technology allows us to determine the kon, koff and KD of interactions, providing deeper insight into binding events compared to other techniques that only give endpoint measurements, such as pull-down assays. SPR is necessary not only for publications but for the advancement of many fields of medicine and medical research as can be seen below with the significant increase in publications that rely on SPR data.
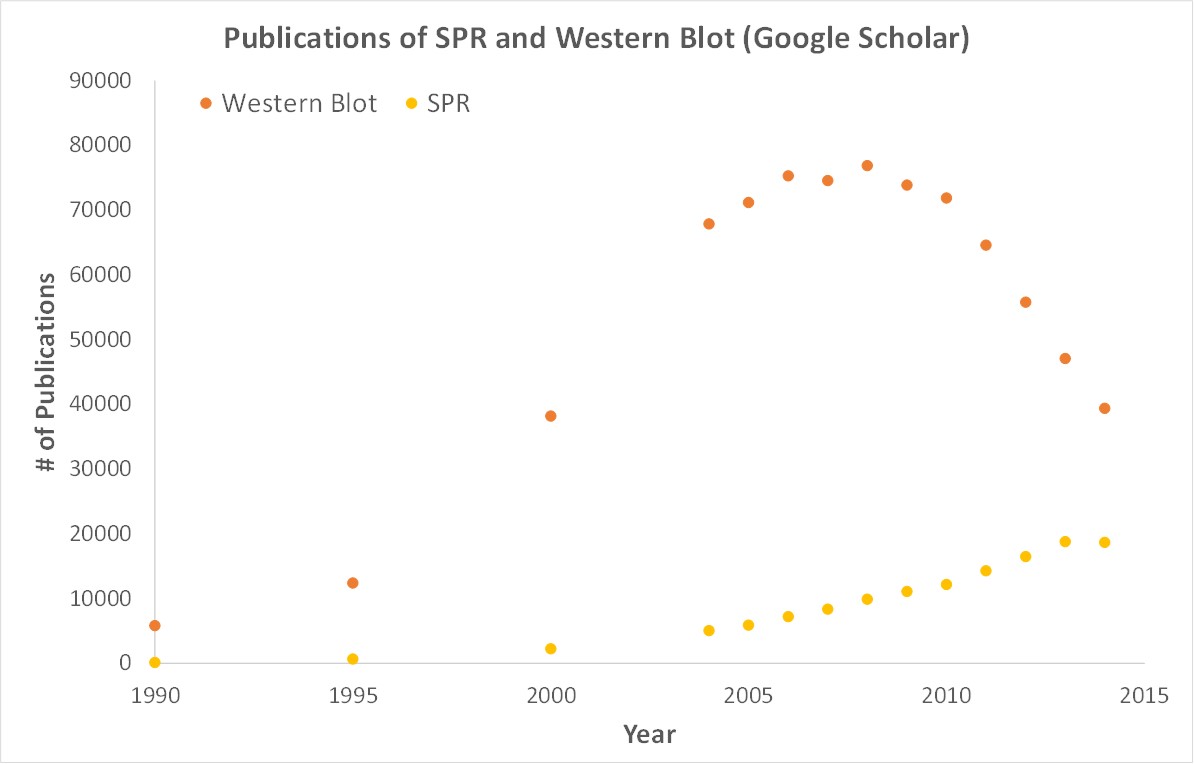
Scientific publications involving SPR have increased drastically over the years. SPR has become fundamental for publications while traditional techniques like Western Blots are becoming less important.
OpenSPR is a user-friendly and low maintenance benchtop SPR solution that is currently being used by hundreds of researchers. With access to SPR technology on your own lab bench you can get the high-quality data you need to accelerate your research and publish faster.
