As the availability of affordable technology increases, reviewers are constantly requesting binding data during the publishing process. However, measuring only affinity or obtaining simplistic yes/no binding data may not satisfy all the requirements you need to successfully publish your research. There are many techniques that dig deeper beneath the surface than just affinity, including fluorescence techniques, which are highly simplistic and low-cost methods for obtaining affinities and/or kinetic constants. However, there are a number of drawbacks when fluorescent techniques are the sole assays employed for obtaining binding information. Here, we will discuss the pros and cons of the most common fluorescence-based binding assays and how you can eliminate the ambiguity associated with these techniques.
Förster Resonance Energy Transfer (FRET)
Pros
– Provides quantitative binding data
– Medium-cost
Cons
– Very limited applications (proteins and nucleic acids only)
– High potential for steric hindrance (10-100 Å distance between interacting partners is required)
– Fluorophores can alter binding properties of interacting partners
– Labour-intensive
Förster resonance energy transfer (FRET) is based on the principle of transferring excitation energy from a donor fluorophore to an acceptor fluorophore. Donor and acceptor fluorophores (ie. blue fluorescent protein and green fluorescent protein, respectively) are fused to the interacting partners and when an interaction between these two proteins occurs, a quenching event can be observed; donor fluorescence intensity decreases while the acceptor fluorophore emission increases simultaneously. In this respect, it is possible to obtain quantitative binding data.
However, it is important to note that since an energy transfer is required for the phenomenon to take place, interacting partners must be within 10-100 Å of one another. When working with very large proteins or large protein complexes, this can become highly problematic. Moreover, since the distance between interacting partners and steric hindrance are important, this will restrict the variety of applications that can be carried out using FRET. Fluorophores can also alter the conformation of your interacting partners and in this respect, compromise the validity of your data.
Fluorescence Polarization (FP)
Pros
– High-throughput
– Real-time data
– Can be used with small molecules
Cons
– Does not provide kinetic constants
– Limited applications (proteins, nucleic acids, small molecules)
– Fluorophores can alter binding properties of interacting partners
– Labour-intensive
– Validating tools are highly recommended
Fluorescence polarization (FP) is based on the principle that when polarized light excites a fluorophore attached to a biomolecule of interest, otherwise known as a “probe” or a “tracer”, light will be emitted in the same planar orientation. Notably, biomolecules rotate and emit in an unlimited amount of configurations, so an excitation filter is employed in FP assays. When light is emitted in the same planar orientation as the filter’s planar orientation, this light will pass through the filter and subsequently, produce a polarization signal. When an interaction between two interacting partners takes place, the molecular weight of the complex increases and the polarization of light is maintained for a longer period of time, which in turn, results in a higher polarization signal.
It should be noted that FP can be used in a high-throughput assay format to determine Kd of proteins, nucleic acids or small molecules but unfortunately, will not provide kinetic constants (on/off rates). Moreover, it is important to cross-reference FP-derived Kd measurements with alternative tools because fluorophores can non-specifically bind to the unlabelled protein binding partner and thus, invalidate your binding data.
Eletrophoretic Mobility Shift Assay (EMSA)
Pros
– Low-cost
– Low-tech
– Non-destructive
Cons
– Does not provide kinetic constants (on/off rates)
– Very limited applications (proteins, nucleic acids)
– Low-throughput
– Fluorophores can alter binding properties of interacting partners
– Labour-intensive
– Validating tools are highly recommended
Eletrophoretic Mobility Shift Assay (EMSA), otherwise known as a gel shift assay, gel mobility shift assay, band shift assay or gel retardation assay, is commonly employed to characterize protein-DNA and protein-RNA interactions. Polyacrylamide or agarose gels electrophoretically separate protein-DNA or protein-RNA complexes of interest from the rest of the mixture based on size and apparent charge. Notably, the DNA or RNA probe(s) of interest (with no protein present; the control lane) will migrate down the gel at a predictable location. However, when the protein of interest is mixed with the DNA or RNA probe and assuming the interacting partners bind, a large complex will be observed and identified by a “shifted” band. This band will reside significantly closer to the loading wells on the gel than the negative control. If the starting concentrations of protein/probe(s) and stoichiometry are known, a Kd value can be determined. It should be noted that for appropriate visualization of the “gel shift”, fluorescent labelling is required, unless radiolabelling is preferred.
Unfortunately, this technique may only be used to measure Kd for protein-nucleic acid interactions, making this technique highly limited in terms of applications. Provided the nature of how EMSA’s are carried out, these assays are highly labour intensive; fluorescent labels (or radiolabels) need to be conjugated on your DNA or RNA probe(s) of interest, polyacrylamide or agarose gels must be prepared, run, visualized and analyzed. Most gels require 1.5 to 2 hours to simply run, not including preparation or analysis time. Moreover, this technique cannot provide quantitative binding kinetics, which reviewers are increasingly seeking from investigators as technologies become more accessible. This publication provides an example of how researchers used EMSA for basic binding data and Surface Plasmon Resonance to validate their findings.
Microscale Thermophoresis (MST)
Pros
– Versatility in applications
– Immobilization-free
Cons
– Does not provide kinetic constants (on/off rates)
– Fluorophores can alter binding properties of interacting partners
– High-cost
Microscale Thermophoresis (MST) is a biophysical technique employed for characterizing affinities of biomolecular interactions. Samples are loaded into a capillary tube and the labelled partner is maintained a constant amount, while the unlabelled partner is loaded at varying concentrations. When an infrared laser is applied to the fluorescent partner, which is usually conjugated with a fluorescent label (unless many tryptophan residues are present), a temperature gradient is established. Subsequently, an LED light is applied to the sample and the fluorescence intensity changes upon excitation of the fluorescent molecule. By plotting calculated values over the log of sample concentration, a Kd can be derived.
Similar to the aforementioned fluorescent techniques, there are some limitations associated with MST. Fluorescent labels can be labour intensive to conjugate to binding partners and may alter the binding properties of your interacting partners (ie. non-specific binding to the fluorescent label). Moreover, MST can be very expensive, especially in a research environment where funding can be limited for individual investigators.
Fluorescence vs. Label-Free Techniques Comparison
After reviewing the pros and cons of fluorescent binding techniques, a few common themes can be observed:
- Cannot directly measure kinetic constants (on/off rates)
- Limited applications
- Fluorophores can alter the binding properties of interacting partners
- Labour-intensive (ie. labelling molecules)
- Validating tools are often required
With that in mind, a label-free method of investigating biomolecular interactions would be advantageous as an alternative or complement to fluorescent techniques. Label-free techniques are less labour-intensive (do not require labels), will provide more accurate binding information (less non-specific binding) and when using the right tool, will provide direct measurement of on/off rates.
Label-Free Techniques: Surface Plasmon Resonance
Surface Plasmon Resonance (SPR) is a label-free technology which allows researchers to directly measure binding constants between two biomolecules. SPR allows you to determine the kon, koff and Kd of interactions, providing deeper insight into binding events compared to other techniques that only provide affinity measurements, such as FRET, FP, EMSA or MST. SPR is necessary not only for publications, but for the advancement of many fields of medicine and medical research; this is illustrated in the figure below, demonstrating the significant increase in publications that rely on SPR data.
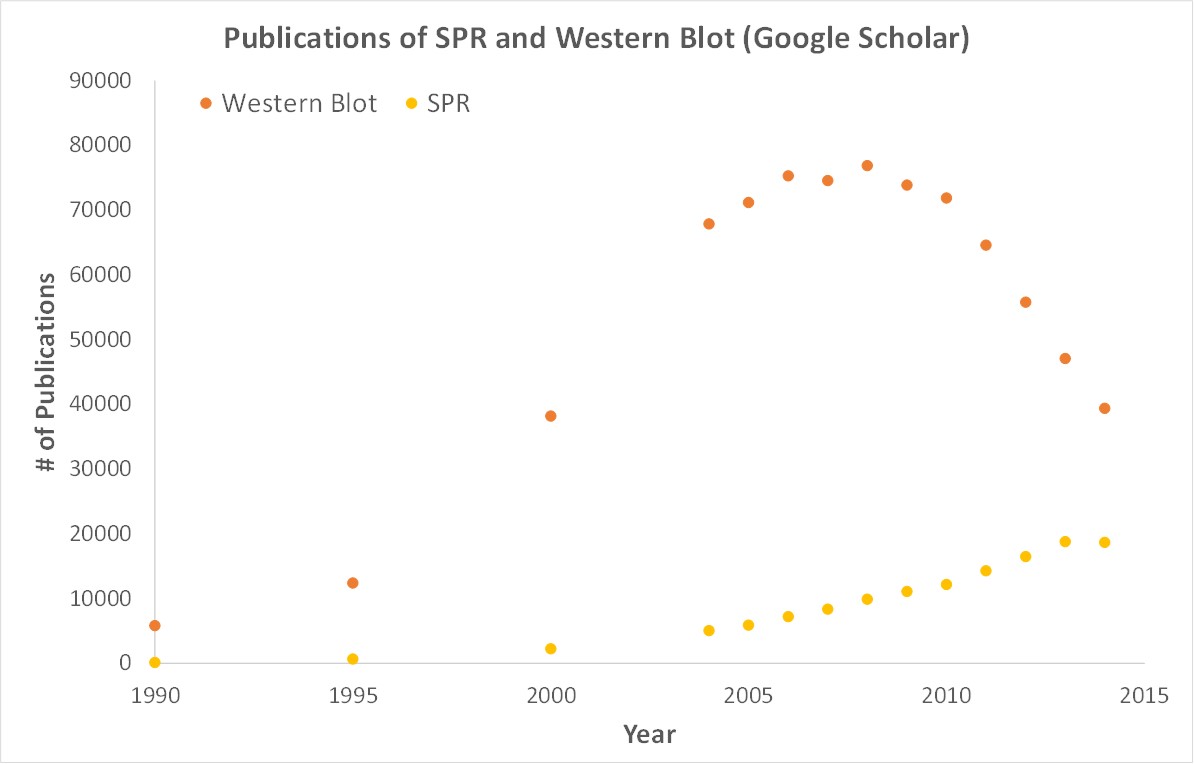
Scientific publications involving SPR have increased drastically over the years. SPR has become fundamental for publications while traditional techniques like Western Blots are becoming less important.
SPR is a label-free technology which allows researchers to quantitatively analyze binding between two biomolecules. SPR technology allows us to determine the kon, koff and KD of interactions, providing deeper insight into binding events compared to other techniques that only give endpoint measurements, such as pull-down assays. SPR is necessary not only for publications but for the advancement of many fields of medicine and medical research as can be seen below with the significant increase in publications that rely on SPR data.
Historically, Surface Plasmon Resonance has been a very necessary yet, inaccessible tool based on the cost of an instrument and the associated consumables. However, there is an affordable option available; OpenSPR is a user-friendly and low-maintenance benchtop SPR solution that is currently being used by hundreds of researchers. With access to SPR technology on your own lab bench you can get the high-quality data you need to accelerate your research and publish faster.
