Virulence factors are tools employed by pathogenic organisms to utilize, evade, colonize, suppress or destroy normal immune responses in a host. An example of a virulence factor is lipid A, a component of endotoxins produced by gram-negative bacteria, such as E. coli. In the context of treating pathogenic infections, virulence factors are the primary targets of antibiotics or anti-retroviral therapies. However, resistance mechanisms in pathogens continue to surface at an alarming rate, largely due to the overuse and misuse of traditional therapies. The problem is so widespread that the World Health Organization (WHO) called on the United Nations General Assembly to commit to responsible administration of antimicrobial therapies in September 2016. In this respect, it’s clear that virulence factors must be understood in more detail when new therapeutic options are inevitably required in the future.
In this new publication, Dr. Judith Kornblatt and Dr. Jack Kornblatt use the OpenSPR’s localized surface plasmon resonance technology to obtain the key binding data needed for their latest discovery. The publication titled, “The influence of truncating the carboxy-terminal amino acid residues of streptococcal enolase on its ability to interact with canine plasminogen” uses binding kinetics data generated from the OpenSPR to assess the capacity of octameric streptococcal enolase to interact with plasminogen, which indicates S. pyogenes infections.
About the Publication
Streptococcus pyogenes is a gram-positive bacterial species and is responsible for infections such as pharyngitis, scarlet fever or impetigo. In more severe instances, S. pyogenes can cause life-threatening infections such as necrotizing fasciitis, otherwise known as flesh-eating disease which destroys soft tissues like skin and fat, and is fatal in 25% of cases.There are a number of virulence factors that contribute to S. pyogenes infections, including binding of enolase to plasminogen.
Streptococcal enolase is a 47 kDa, homo-octameric enzyme which exists as a tetramer of dimers. Enolase is typically responsible for the second last step in the glycolytic pathway, but also maintains important binding properties in the context of S. pyogenes infections. Moreover, plasminogen, the inactive precursor of plasmin found in blood, is a protease responsible for the catalysis of blood clots.
Historically, the interaction between these two proteins in their native states has not been described as strong. However, it has been shown that when the octameric structure of enolase is disrupted, the interaction between these proteins becomes remarkably robust. Notably, activation to plasmin can occur when enolase and plasminogen interact. When this event occurs, plasmin will begin to degrade proteinaceous components of tight junctions and subsequently encourages the pathogen to spread. This is an effective virulence factor of S. pyogenes and may contribute to flesh-eating disease. It is quite important to understand this crucial interaction and in order to explore the kinetic parameters, the investigators used surface plasmon resonance.
Why was OpenSPR instrumental for this research?
Surface plasmon resonance (SPR) was used to quantitatively characterize the interaction between truncated Streptococcal enolase and canine plasminogen. Truncated Streptococcal enolase samples were N-terminally histidine tagged and immobilized on an NTA sensor chip; notably, 4 amino acid residues were removed from this immobilized enolase sample. Next, canine plasminogen was injected in duplicates and in concentrations ranging from 0.11 to 9.1 µM and binding kinetics were measured with very accurate fits. Upon completion of their binding studies, the investigators concluded that truncated species of Streptococcal enolase strongly interact with canine plasminogen and that two binding sites are involved. The results provided a significantly deeper understanding of S. pyogenes virulence factors and may allow future studies to be focussed on novel therapeutic interventions.
Why is SPR critical for publications? How does OpenSPR help?
SPR is a label-free technology which allows researchers to quantitatively analyze binding between two biomolecules. SPR technology allows us to determine the kon, koff and KD of interactions, providing deeper insight into binding events compared to other techniques that only give endpoint measurements, such as pull-down assays. SPR is necessary not only for publications but for the advancement of many fields of medicine and medical research as can be seen below with the significant increase in publications that rely on SPR data.
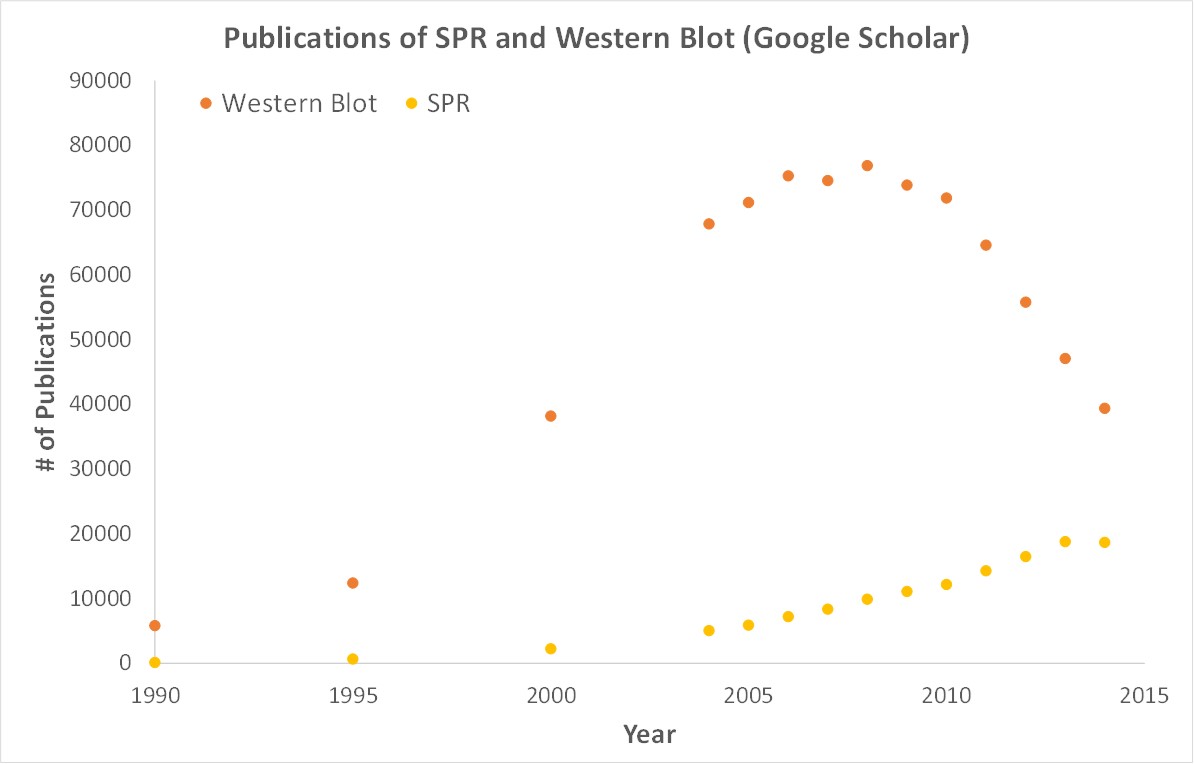
Scientific publications involving SPR have increased drastically over the years. SPR has become fundamental for publications while traditional techniques like Western Blots are becoming less important.
OpenSPR is a user-friendly and low maintenance benchtop SPR solution that is currently being used by hundreds of researchers. With access to SPR technology on your own lab bench you can get the high-quality data you need to accelerate your research and publish faster.
