Galvanize your productivity with Alto’s epitope binning application:
Pre-designed, intuitive, and sample-friendly.
Early antibody discovery involves the screening of a large number of molecules to identify hits and narrow them down to more promising leads. Efficient epitope binning plays a key role in identifying high quality leads to inform the downstream development process, and label-free biosensors have emerged as a powerful tool for this application. Despite significant advances in label-free tools for antibody discovery, there remains a need for more powerful, high-throughput alternatives to streamline the process without compromising on data quality.
Powered by digital microfluidics, Alto™ revolutionizes real-time interaction analysis by reducing the cost and complexity of antibody discovery. Our enhanced epitope binning module streamlines experimental workflow, empowering you with high-quality data and giving you back precious time and resources.
Continue reading below to learn how you can galvanize your antibody discovery with Alto’s enhanced binning module!
Table of contents
- Epitope characterization in antibody discovery
- Basics of a competition assay
- Approaches for epitope binning
- Epitope binning with Alto™
- Conclusion
1. Epitope characterization in antibody discovery
Therapeutic antibodies are an important and growing part of our disease treating arsenal, providing more efficient and specific alternatives to conventional drugs. Once administered, therapeutic antibodies work by directly neutralizing disease-causing agents, blocking cellular interactions, stimulating immune responses, or delivering therapeutic payloads to specific locations. They have shown great potential in treating a wide range of conditions, including cancers, autoimmune, and infectious diseases.
Antibodies orchestrate immune responses by recognizing and docking to epitopes on other molecules, a mechanism that can be leveraged for designing therapeutics. The first step in antibody discovery is to identify and validate a disease target that can be used to elicit a therapeutic effect. Then the work begins to find antibodies that specifically dock to epitopes on the target of interest. One approach taken is epitope binning, the process of screening and categorizing antibodies based on their epitope binding properties. Efficient and accurate epitope binning is important throughout the therapeutic development process, but is especially key in:
- Early antibody discovery: Because early antibody discovery generates hundreds of ‘hits’, narrowing them down to more promising ‘leads’ becomes a challenge. Almost 90% of drugs entering clinical Phase I do not reach commercialization, indicating shortfalls in the lead-to-hit phase. With only 1 out of 10 drug candidates passing regulatory approval, eliminating low-quality leads as early as possible is important in improving the efficacy and lowering the overall cost of drug development.1
- Immunogenicity testing: Characterizing immunogenicity, or patient response to antibody drugs by characterizing anti-drug antibodies (ADAs), is an important part of assessing drug safety in clinical trials. More recently, there has been a call for earlier epitope characterization of ADAs to avoid downstream immune responses against leads.2
Developing high-throughput analytical tools enabling better informed antibody selection is imperative to devising the next generation of therapeutics. High-throughput label-free biosensors have emerged to fit this need by providing a platform for fast and accurate epitope characterization.
2. Basics of a competition assay
A competition assay or a competitive ligand binding assay (LBA) is an assay wherein two molecules compete for the same binding site, which can help elucidate binding properties and mechanisms of action.
Epitope binning employs a competitive assay to characterize the simultaneous binding of monoclonal antibodies (mAbs) to an antigen, tested in a pairwise manner, for determining whether they block one another’s binding to the same epitope. If the binding of one mAb to the antigen prevents the binding of the other, both mAbs can be clustered into groups (or bins) to indicate they compete for the same or similar epitope. If both mAbs are able to bind to the antigen, then they are considered to bind to distinct, non-overlapping epitopes.
3. Approaches for epitope binning
Traditional techniques such as enzyme-linked immunosorbent assays (ELISA) and western blots (WB) require significant amounts of sample, time-consuming washing and incubation steps, and depend on the use of tags for performing competition assays. In contrast, label-free biosensors such as BLI and SPR have allowed for more automation and throughput in facilitating and analyzing epitope binning experiments. But they aren’t without their limitations.
Traditional SPR instruments often have a large footprint and incorporate complex fluidic systems that are cumbersome to maintain. BLI instruments tend to consume a large amount of sample in epitope binning applications due to the need to load in 96- or 384-well plates. In requiring multiple wells of solution antibody and antigen per run, a full pairwise analysis can consume upwards of 100 µg per antigen.
By integrating digital microfluidics, our Digital SPR platform, Alto™, builds on the capabilities of conventional SPR platforms to enhance competition assays and epitope characterization studies.
4. Epitope binning with Alto™
Powered by digital microfluidics, Alto™ revolutionizes real-time SPR interaction analysis by streamlining your assays while providing publication-quality data (Figure 1). Its intuitive interface and user-friendly software, the Nicosystem™, take you from assay design to data analysis within a single day.
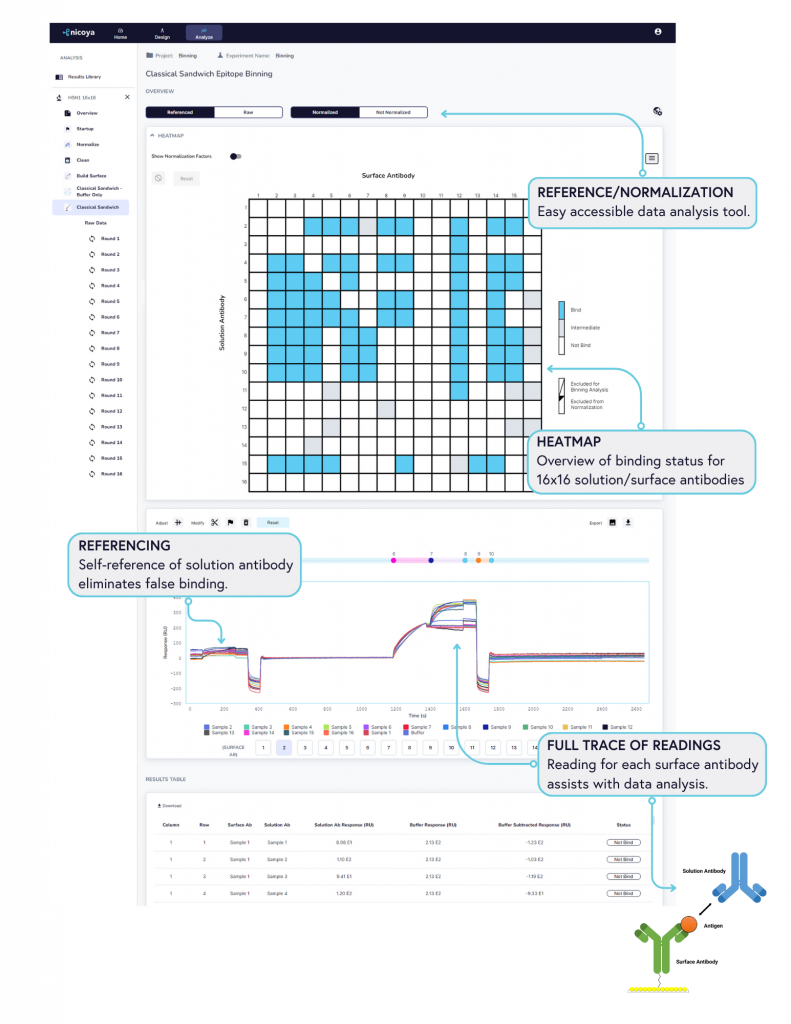
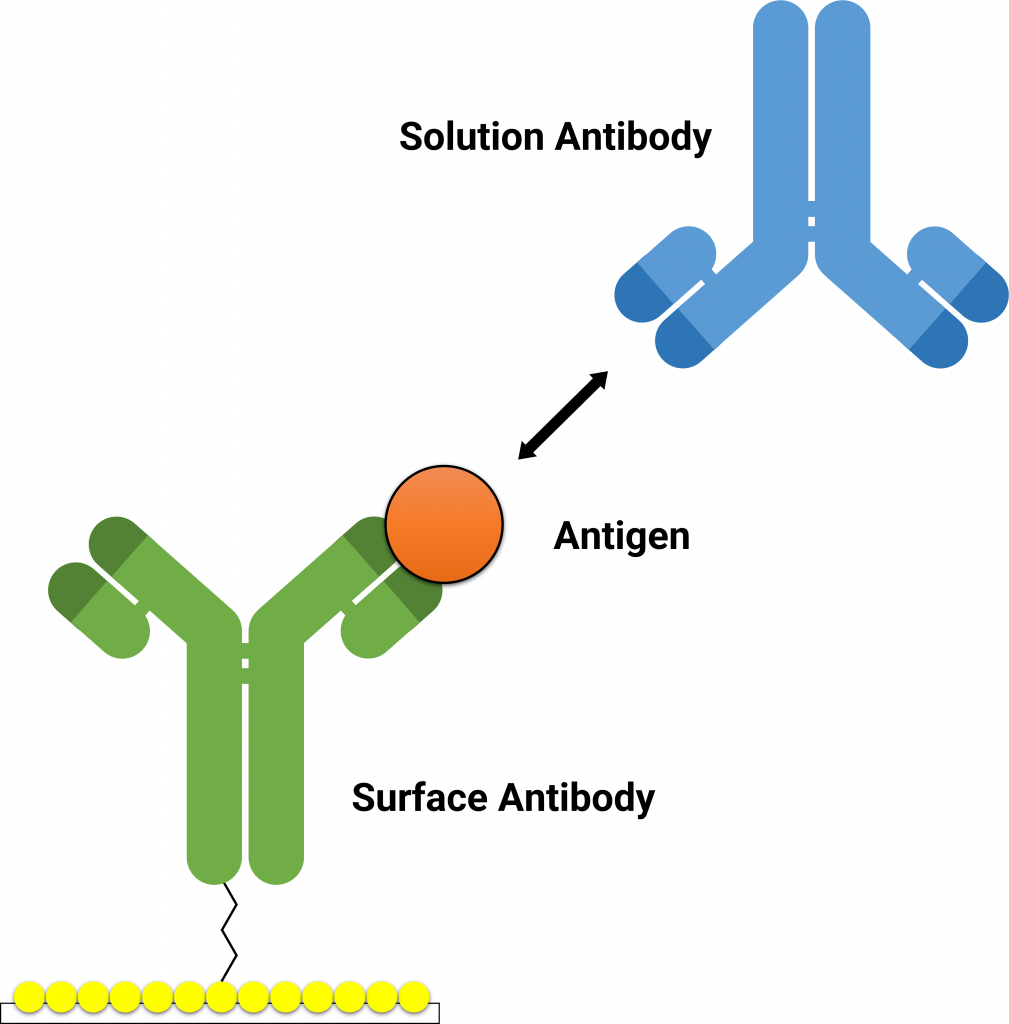
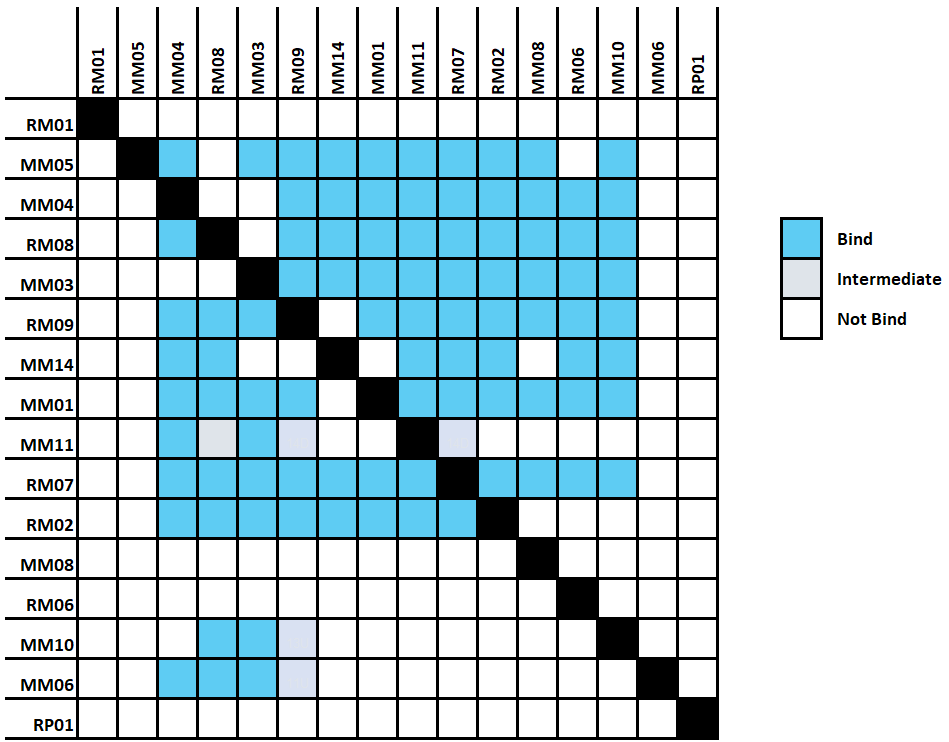
The Alto™ epitope binning protocol utilizes a classical sandwich format (Figure 2). In the classical sandwich assay, the first mAb (surface antibody) is immobilized onto the sensor, followed by injection of the antigen, and finally the second mAb (solution antibody). The binding data of all antibodies are then used to generate heatmaps by grouping binders and non-binders (Figure 3).
Designing your experiment
Alto’s enhanced binning application enables the simultaneous analysis of 256 interactions by collapsing 16×16 epitope binning experiments into single runs that can be both designed and analyzed within a day.
One major advantage of performing epitope bins on Alto™ is its ultra-low sample consumption, requiring a total of 100 ng per antibody and ~1 μg of antigen for a 16×16 run.
As well, in contrast to the high learning curves of SPR systems, Alto’s pre-designed, automated assays simplify competition assays to make epitope binning more accessible to a wide range of users. Instead of having to build each experimental protocol from scratch, Alto™ users simply select the epitope binning protocol and either enter or import their sample information (in CSV format) once. The protocol will self-populate with the samples provided and a color-coded plate map will be generated to guide sample loading at the bench. Adjustments can also be easily made to contact and interaction times before saving the protocol and pushing it to the device to run.
Analyzing your data
With its seamless user interface, Alto™ enables researchers to go from assay design to data analysis in just one click. Figure 4 illustrates some of the key tools and accommodations we’ve built into Alto’s analysis software, the Nicosystem™, to simplify post-processing of epitope binning experiments:
- Heat map: All binding data are collapsed into heatmaps, providing clear visual information on the binding status of antibodies (bind/intermediate/no bind level). Hovering on the heatmap reveals individual antibody sample names for easy overview.
- Full trace sensorgram: Full trace sensorgrams are generated to provide insight into antibody-antigen-antibody interactions, enabling users to investigate the binding curve with all the information.
- Normalization and reference: This provides flexibility for investigating the big data size without the use of third-party analysis methods, a feature unique to Alto™, as other systems require additional software packages to analyze binning data.
- Referencing: As a powerful built-in tool, self-referencing of solution antibody eliminates false binding, providing you with data you can trust.
- Result table: Complete information of all 256 samples is captured in the result table, providing data complimentary to the heat map for easy analysis.
5. Conclusion
Therapeutic antibodies are treating diseases with more specificity and efficiency than ever before. Maximizing their potential hinges on the continued development of analytical tools to improve the characterization of high quality leads and fail low-quality ones earlier in the pipeline. Biosensor-based epitope characterization has emerged as a powerful tool in this regard, providing a high-throughput, label-free platform for antibody-based drug discovery.
Our next-gen digital SPR platform Alto™ builds on this through its revolutionary integration of DMF, thereby automating and miniaturizing epitope binning assays, and ultimately reducing the cost and complexity of antibody discovery. For those interested in earlier results or other non-binning applications, Alto™ offers crude sample compatibility and numerous other applications such as library screening and kinetics characterization. To learn more, chat with our team of Application Scientists today!
Interested in learning more about Alto™ Digital SPR? Book your demo today!
References
- Hay M, Thomas DW, Craighead JL, Economides C, Rosenthal J. Clinical development success rates for investigational drugs. Nat Biotechnol. 2014;32(1):40-51. doi: 10.1038/nbt.2786
- McMaster M, Mohr K, Page A, Closmore A, Towne F, Brooks BD. Epitope Characterization of Anti-drug Antibodies—A Tool for Discovery and Health: An overview of the necessity of early epitope characterization to avoid anti-drug antibodies and promote patient health. Expert Opin Biol Ther. 2021;21(6):705-715. doi: 10.1080/14712598.2021.1863942