Hepatitis B virus (HBV) is linked to more than 800,000 deaths per year worldwide. It remains a global health concern in many developing regions such as Asia and sub-Saharan Africa. HBV tends to manifest and replicate itself in human hepatic cells and is primarily transmitted vertically, from mother to offspring. Furthermore, exposure to infected bodily fluids (ie. sharing needles, sexual intercourse with an infected person) can result in horizontal transmission.
HBV is a double stranded DNA (dsDNA) virus belonging to the Hepadnaviridae viral family and has been vaccinable for almost four decades. However, conventional HBV vaccination strategies rely on intramuscular administration of the small hepatitis B surface (SHBs) protein produced from transfection in S. cerevisiae cells. It is important to note that SHBs is only one of three main therapeutic HBV antigens and literature suggests that targeting the remaining two epitopes, namely the pre-S1 and and pre-S2 epitope, may confer an efficacious advantage.
In a recent publication, Dr. Yu-Ching Lee uses the OpenSPR’s localized surface plasmon resonance technology to generate the key binding data needed for their discovery. The publication “Interaction of S17 antibody with the functional binding region of the hepatitis B virus pre-S2 epitope” uses binding data generated with the OpenSPR to probe the interactions between single-chain variable fragment (scFv) antibodies and the pre-S2 epitope, which may augment existing HBV immunotherapeutic strategies.
About the Publication
The HBV consists of three major viral envelope proteins termed the hepatitis B surface proteins (HBs), which consists of the 226 amino acid residue small hepatitis B surface protein (SHBs), the 281 residue middle hepatitis B surface protein (MHBs), and the 400 residue large hepatitis B surface protein (LHBs). The traditional vaccination strategy, developed in the 1980’s, has revolved around the intramuscular administration of SHBs in which transfected yeast cells are the manufacturer of this antigen, which is subsequently formulated into commercial immunotherapeutics. However, the frequency of non-responders upon vaccination is high and this is why a need has emerged for more effective immunoprotection methods.
Literature has shown that employing MHBs and LHBs (pre-S1 and pre-S2 epitopes, respectively) in immunotherapeutic formulations has proven to be an efficacious, preventative vaccination strategy in murine models. Previous studies have also indicated that introduction of the pre-S/S epitopes may also provide an effective intervention for more sustained HBV infections. Specifically, the pre-S2 epitope has been implicated as a major mediator in attachment of HBV to hepatocytes, a hallmark of HB viral invasion. It is noteworthy to mention that patient exposure to the pre-S2 epitope elicits a significantly higher immune response than other aforementioned epitopes; residues 123-145 of this polypeptide are notorious to be associated with effective neutralization. In this way, the interaction of the pre-S2 epitope with engineered antibodies has become a popular domain of study for immunologists.
In this publication, the researchers employed phage-display techniques to isolate human scFvs from an HBV patient and subsequently selected leading candidates through ELISA and Western Blots for further analyses against the pre-S2 epitope. Following flow cytometry and immunocytochemical staining the scFv antibody, S17 was conscripted for computational modelling and surface plasmon resonance (SPR) analysis.
Results from surface plasmon resonance studies suggest that scFv S17 was a compelling neutralizing agent against the pre-S2 epitope found on the HBV viral envelope. It should be noted that binding of the complete IgG molecular structure may provide an even higher affinity.
Why was OpenSPR instrumental for this research?
Surface plasmon resonance (SPR) was used to determine the interactions between the pre-S2 epitope and the scFv antibody, S17. The pre-S2 peptide was immobilized on a streptavidin sensor and the antibody-antigen experiment was run on the OpenSPR system. By using OpenSPR, the authors were able to determine the kinetic data (association and dissociation rate constants, equilibrium constant, and affinity) of the interaction between the S17 antibody and immobilized pre-S2 epitope. SPR binding curves showed S17 was efficacious in interacting with pre-S2 and the quantitative data from this experiment manifested an equilibrium dissociation constant (KD) of 4.2 x 10-8 M. Results generated from this study indicate that the scFv S17 antibody may serve as an effective immunotherapeutic agent by targeting the HBV pre-S2 epitope and could translate into an improved vaccination strategy against HBV infection. By using the OpenSPR, these researchers were able to get SPR data from their own bench which helped them accelerate their research and publish their discovery faster.
Why is SPR critical for publications? How does OpenSPR help?
SPR is a label-free technology which allows researchers to quantitatively analyze binding between two biomolecules. SPR technology allows us to determine the kon, koff and KD of interactions, providing deeper insight into binding events compared to other techniques that only give endpoint measurements, such as pull-down assays. SPR is necessary not only for publications but for the advancement of many fields of medicine and medical research as can be seen below with the significant increase in publications that rely on SPR data.
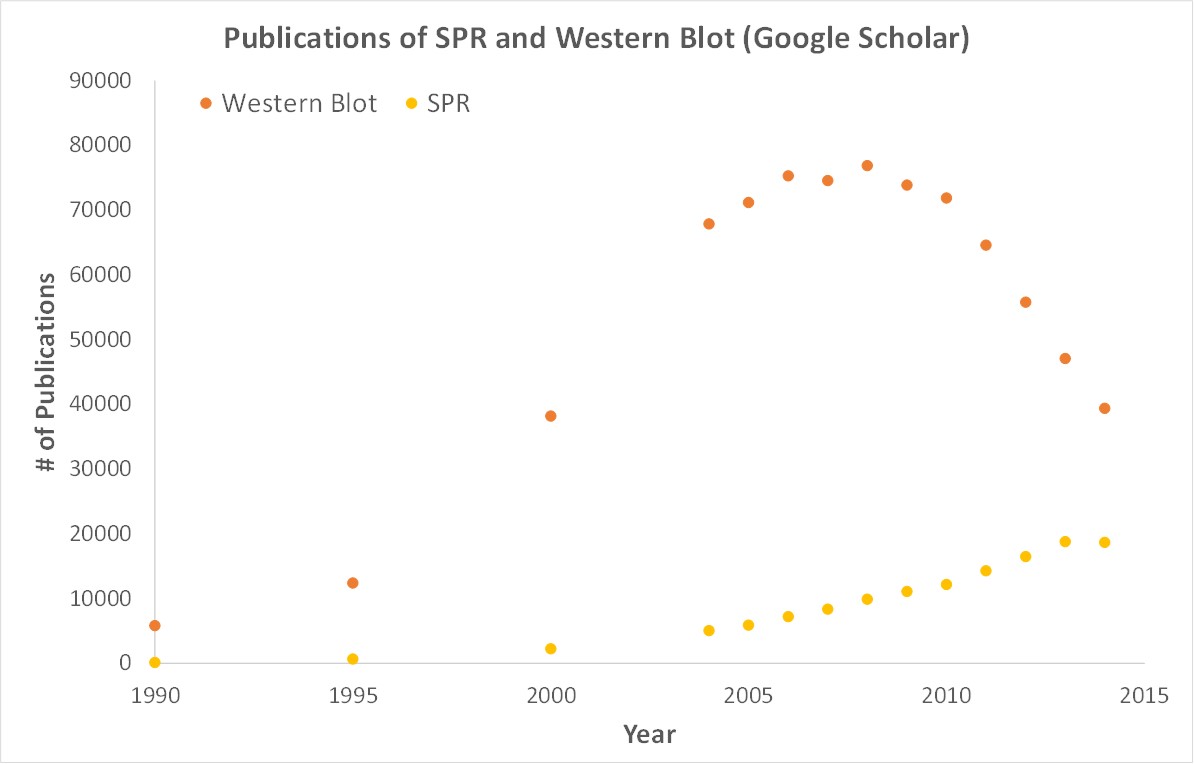
Scientific publications involving SPR have increased drastically over the years. SPR has become fundamental for publications while traditional techniques like Western Blots are becoming less important.
OpenSPR is a user-friendly and low maintenance benchtop SPR solution that is currently being used by hundreds of researchers. With access to SPR technology on your own lab bench you can get the high-quality data you need to accelerate your research.
[1] Budkowska, A., Dubreuil, P., & Pillot, J. (1987). Detection of antibodies to pre-S2 encoded epitopes of hepatitis B virus by monoclonal antibody-enzyme immunoassay. Journal of Immunological Methods,102(1), 85-92. doi:10.1016/s0022-1759(87)80013-3
[2] Chang, C., Chang, F., Chiang, C., Lo, Y., Lin, T., Chen, W., . . . Lee, Y. (2018). Interaction of S17 Antibody with the Functional Binding Region of the Hepatitis B Virus Pre-S2 Epitope. Viral Immunology. doi:10.1089/vim.2017.0200
[3] Locarnini, S. (2004). Molecular Virology of Hepatitis B Virus. Seminars in Liver Disease,24, 3-10. doi:10.1055/s-2004-828672
[4] Shouval, D., Ilan, Y., Adler, R., Deepen, R., Panet, A., Even-Chen, Z., . . . Gerlich, W. (1994). Improved immunogenicity in mice of a mammalian cell-derived recombinant hepatitis B vaccine containing pre-S1 and pre-S2 antigens as compared with conventional yeast-derived vaccines. Vaccine,12(15), 1453-1459. doi:10.1016/0264-410x(94)90155-4
[5] Shouval, D., Roggendorf, H., & Roggendorf, M. (2015). Enhanced immune response to hepatitis B vaccination through immunization with a Pre-S1/Pre-S2/S Vaccine. Medical Microbiology and Immunology,204(1), 57-68. doi:10.1007/s00430-014-0374-x
[6] World Health Organization. (2018, July 18). Hepatitis B. Retrieved from http://www.who.int/news-room/fact-sheets/detail/hepatitis-b
