Are you missing quantitative binding data in your publication? Dr. Wright, Assistant Professor at James Madison University, uses OpenSPR’s surface plasmon resonance technology to provide him with the key binding data needed for his recent publication. The research paper “A novel FLNC frameshift and an OBSCN variant in a family with distal muscular dystrophy” published in PLOS One (Public Library of Science) uses SPR data generated with OpenSPR to study the genetic mutational links to muscular protein function with titin, the z-disc, and obscurin and how these impact muscular dystrophy disease penetrance. With SPR data, Dr. Wright was able to publish an extensive study on muscular dystrophy. Continue reading to learn more about this discovery-based research and how SPR data validated the effects of the Z-disc mutation and provided quantitative affinity and kinetic data.
Understanding Muscular Dystrophy Protein Function
This publication advances muscular dystrophy research by examining the genetic mutational links to muscular protein function with titin, the z-disc, and obscurin and how these impact disease penetrance. The interaction between the obscurin Ig58/Ig59 and the titin ZIg9/ZIg10 is important for targeting obscurin to the Z-disk. A novel FLNC frameshift and OBSCN variant mutation was identified in members of a French family affected by distal myopathy. Furthermore, the p.Arg4444Trp variant is localized within the OBSCN Ig59 domain that, together with Ig58, binds to the ZIg9/ZIg10 domains of titin at Z-disks. To verify the effect of the OBSCN variant on the interaction with titin, SPR experiments were performed to determine the binding kinetics of mutant and wild type Ig58/Ig59 domains to the ZIg9/ZIg10 domains of titin. The protein-protein interaction studies were run using Nicoya’s OpenSPR instrument and GST sensor chip.
Why was OpenSPR instrumental for this research?
The SPR studies showed Ig58/Ig59 and titin ZIg9ZIg10 interaction binding kinetics – giving more detail into the interaction by providing on and off rates. The results from the SPR experiments indicated that the OBSCN variant results in a significant decrease in binding between the Ig58/Ig59 domains and the titin ZIg9/ZIg10. Furthermore, there was a ~15-fold weaker interaction between titin and the mutant obscurin Ig58/Ig59 region, with a KD of 1.6 μM and 108 nM for the mutant and the wild type Ig58/Ig59 domains, respectively. With these results, Dr. Wright and his colleagues were able to show that SPR data validated the effects of the Z-disc mutation and provided quantitative (15 fold decrease) affinity and kinetic data. In addition, OpenSPR allowed Dr. Wright and his research lab to obtain SPR data from their own bench.
Why is SPR critical for publications? How does OpenSPR help?
SPR is a label-free technology which allows researchers to quantitatively analyze binding between two biomolecules. SPR technology allows us to determine the kon, koff and KD of interactions, providing deeper insight into binding events compared to other techniques that only give endpoint measurements, such as pull-down assays. SPR is necessary not only for publications but for the advancement of many fields of medicine and medical research as can be seen below with the significant increase in publications that rely on SPR data.
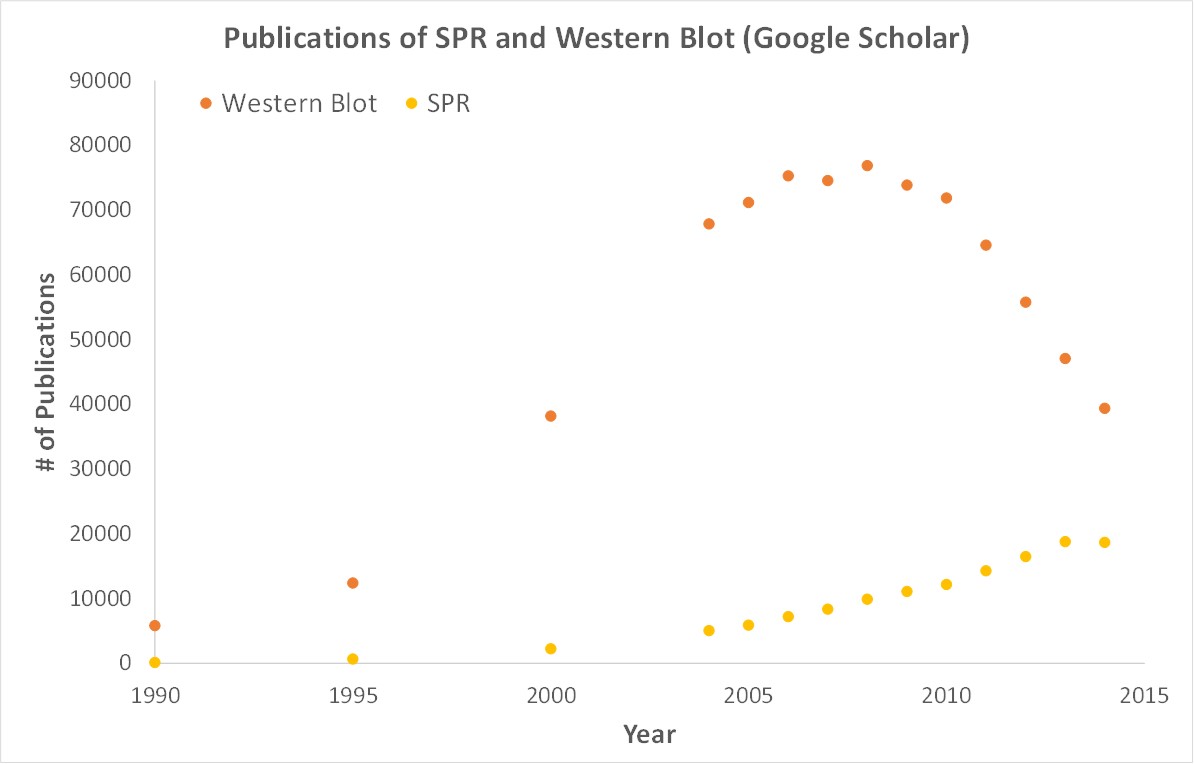
Scientific publications involving SPR have increased drastically over the years. SPR has become fundamental for publications while traditional techniques like Western Blots are becoming less important.
OpenSPR is a user-friendly and low maintenance benchtop SPR solution that is currently being used by hundreds of researchers. With access to SPR technology on your own lab bench you can get the high quality data you need to accelerate your research and publish faster.
