Virus-like particles (VLPs) are complex molecules composed of multiple protein subunits that resemble a specific virus without its genetic material. VLPs allow researchers to safely study viruses without running the risk of exposure to a viral infection. Compared to other compounds typically used to study viruses (such as epitopes subjected to inactivation treatments), VLPs represent conformational epitopes that better resemble the native virus.1 For this reason, VLPs are commonly used in the development of vaccines and other immunotherapies. Virus-like particles have already been crucial in developing a number of vaccines against hepatitis B virus (HBV) and human papillomavirus (HPV).1
As with any therapeutic discovery workflow, screening antibodies for appropriate binding (in this case to a VLP) is a crucial step in development. At this stage, a high-throughput instrument is often used, such as a digital microfluidics-powered SPR instrument. However, virus-like particles are very large, complex molecules composed of multiple protein subunits. As a result, the effective molar concentration of a VLP is low while the analyte surface available for binding (and the resulting stoichiometry for the antibody) is high. This makes kinetic studies challenging, as the overall avidity of a VLP binding system is often unknown. Binding events can also be difficult to measure when an analyte has a molecular weight on the order of several MDa.
Understanding the importance of studying VLP binding in vaccine development, we designed the OpenSPRTM to be capable of detecting large, complex molecules – in addition to a diverse range of other biomolecules. In our latest application note, the binding of a virus-like particle to a captured antibody is studied using the OpenSPR-XTTM instrument. The experiment and findings are summarized below.
Kinetic analysis of a virus-like particle with a captured antibody
In our latest application note, the OpenSPR-XTTM was set up using a PBS-T/sucrose running buffer. A Protein A Sensor was then prepared, and the antibody was captured on the Protein A surface. The binding of the VLP to the antibody was then analyzed over four concentrations of the VLP, regenerating the Protein A surface with Glycine-HCl between injections. The resulting binding curves are shown in Figure 1:
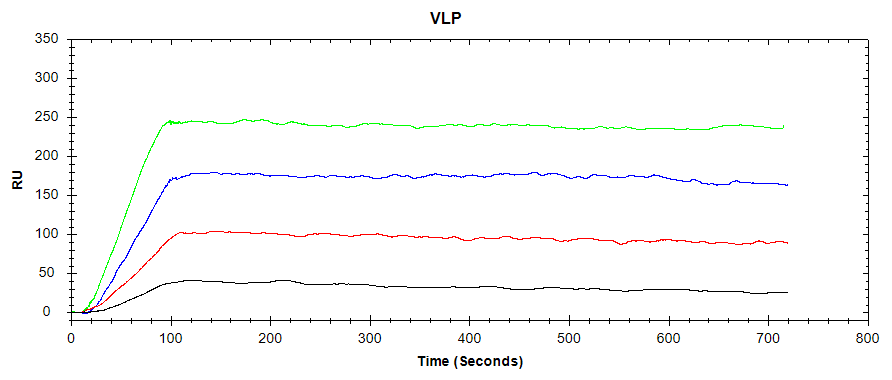
While the avidity of the binding system between the virus-like particle and the captured antibody is unknown due to the complexity of the large VLP, a high-affinity interaction can still be observed. Using sub-nanomolar concentrations of VLP, a clear association phase can be discerned, with virtually no dissociation into the buffer. From this, it can be concluded that the chosen antibody is an excellent candidate to bind this VLP. In a vaccine development workflow, this antibody may then progress to in-vivo testing.
Successful detection of a 10MDa virus-like particle using the OpenSPR-XTTM
Using the OpenSPR-XTTM, researchers successfully detected the binding of a captured antibody to a virus-like particle with an approximate hydrodynamic radius of 40 nm and molecular weight on the order of several MDa. In addition to a diverse range of other biomolecular interactions, the OpenSPRTM is, therefore, an optimal choice for studying VLPs and other large, complex molecules. With two 96-well plates to pull from, the OpenSPR-XTTM offers 24 hours of automated runtime with precise injections (if you’re looking for end-to-end experiment automation, check out AltoTM – the world’s first digital, high-throughput, benchtop SPR system). The intuitive design and affordable cost of the OpenSPRTM system gives any researcher the ability to study quantitative binding kinetics on their own lab bench.
Successfully measure the binding of large & complex molecules with OpenSPR-XT™
DOWNLOAD BROCHUREReferences
- Roldão, A., Mellado, M. C., Castilho, L. R., Carrondo, M. J., Alves, P. M. (2010). Virus-like particles in vaccine development. Expert Rev. Vaccines, 9(10), pp. 1149–1176. Doi: 10.1586/erv.10.115.
