In the arena of drug discovery, there are a number of tools available for researcher’s to characterize binding behaviours of low molecular weight compounds with their target(s). These tools may include fluorescence or radiolabelling assays, keeping in mind that it is always recommended to avoid using labels as these may alter the spin (i.e. this is important when using fluorescence polarization) and may be significantly more hazardous to carry out (i.e. radiation). However, most small molecule binding assays can present their own unique challenges. For example, interactions between proteins and extremely low molecular weight compounds (<200 Da) may not be adequately resolved by surface plasmon resonance (SPR) in a direct kinetic binding assay. If immobilization of the smaller molecular weight binding partner is not feasible, there is a robust and simplistic SPR binding assay that researchers can employ to obtain a KD constant for their small molecules of interest.
What is a competitive ligand binding assay?
A competitive ligand binding assay or competition assay is an indirect quantitative measurement of KD observed when titration of a molecule of interest inhibits binding of the primary target with a known third-party binder. For a simplistic overview of how these molecules interact with one another in the context of a solution competition assay (explained in the next paragraph), here is an example:
A = primary target (protein A)
B = molecule of interest (drug compound B)
C = known third-party binder (protein C)
A binds B
A binds C
B does not bind C
Two Types of Competition Assays
It is important to note that two classes of competition assays exist; a solution competition assay and surface competition assay.
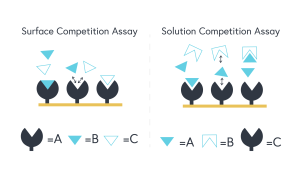
In solution competition, C (the known third-party binder), will be found on the sensor surface and A (primary target) and B (molecule of interest) will be found in the running buffer. When B is titrated at increasing concentrations, lower response values will be observed between A and C. Subsequent mathematical equations can derive an indirectly measured KD constant between A and B.
In surface competition, A (the primary target) will be found on the sensor surface and B (molecule of interest) and C (the known third-party binder) will be found in the running buffer. The same aforementioned themes will apply in this case but it is important to note that when conducting a surface competition assay, C must be substantially larger in molecular weight than B.
The important takeaway between these assay classes is that in solution competition, A (the primary target) can be found in the running buffer, whereas in surface competition, A will be found immobilized on the sensor chip surface. Figure 1 (below) provides an excellent schematic representation of the fundamental differences between each competition assay class.
Figure 1 – The left diagram represents a surface competition assay. The right diagram represents a solution competition assay.
How to Setup a Competition Assay
There are a few experimental parameters to consider when designing a competition assay:
- Do I have an expected KD value between A and B?
- Do I have a known third-party binder (C)?
- Do I have an expected KD value between A and C?
- What is the size difference between B and C?
- Is the size difference large enough between B and C?
- Should I use a solution or surface competition assay?
Once these questions are addressed, you should have a clear indication as to which competition assay class will be most suitable for your application(s). For our example, we will focus on how to conduct a solution competition assay, in very modest terms:
- Using the correct sensor surface chemistry, immobilize C to the sensor
- Starting with the lowest concentration, make 3-5 injections of increasing concentrations of A
- Process binding data with post-processing software to obtain binding constants
- While keeping C immobilized to the sensor, make 3-5 injections of static concentrations of A and increasing concentrations of B (starting with the lowest concentration)
- A decreasing signal should be observed with each injection of increased concentrations of B
- Process the binding data and analyze as a competition assay with post-processing software to obtain a relative KD
Note that the analysis calculations will vary depending on what post-processing software you are using, but this should provide an excellent starting point to understanding competition assays with SPR. At Nicoya, our mission is to improve human life by helping scientists succeed. We have an excellent customer support team that will always be there to help you, from designing your experiment to publishing. Competition assays are an extremely handy tool in drug discovery and can be used in tandem with other technologies for strengthening and validation purposes. However, these technologies typically use a lot of sample, require labels, do not provide enough information and/or require large capital expenditure. Surface Plasmon Resonance (SPR) has been the gold standard for obtaining quantitative binding kinetics data for a number of decades.
Importantly, OpenSPR is a user-friendly and low maintenance benchtop SPR solution that is currently being used by hundreds of researchers. With access to SPR technology on your own lab bench, you can get the high-quality data you need to accelerate your research and publish faster. SPR is necessary not only for publications, but for the advancement of many fields of medicine and medical research as can be seen below with the significant increase in publications that rely on SPR data.
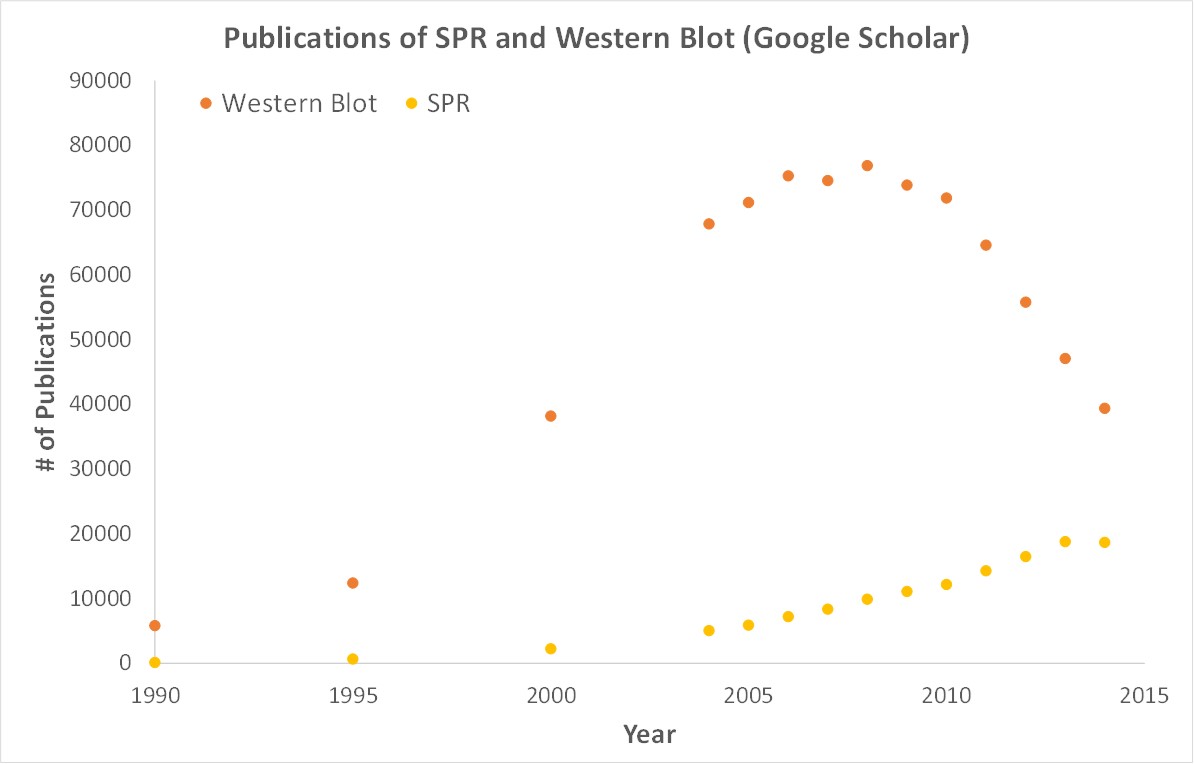
Figure 3. Graph showing the increase in use of SPR in publications.
Resources
- Gesellchen, F., Zimmermann, B., & Herberg, F. W. (2005). Direct Optical Detection of Protein–Ligand Interactions. Methods in Molecular Biology, 305, 17-45. doi:10.1385/1-59259-912-5:017