Nanoparticles (NPs) have been long described as effective vehicles for drug delivery and disease diagnosis. However, upon the introduction of NPs into a biological system, soluble proteins tend to indiscriminately adsorb quite rapidly. Subsequently, these proteins form a corona around the surface of ligand-modified NPs and it is important to note that the formation of these protein corona may intensely alter the binding properties. Consequently, this reduces the binding efficiency and efficacy of the therapeutic vehicle. In this respect, binding efficiency and efficacy of a loaded NP vehicle with and without the presence of protein corona, are very important parameters to consider. In this new publication, Dr. Huile Gao et al. use OpenSPR’s localized surface plasmon resonance technology to provide them with the key binding data needed for their latest discovery on drug delivery mechanisms . The publication titled, “Ligand size and conformation affect the behavior of nanoparticles coated with in vitro and in vivo protein corona” uses binding data generated from the OpenSPR to observe the binding behaviours of ligand-modified NPs possessing protein corona.
About the Publication
Transferrin receptor (TfR) is a transmembrane glycoprotein expressed on the surface of all cells and is primarily responsible for mediating iron transport via it’s iron-binding ligand, transferrin. Importantly, TfR tends to be overexpressed on the surface of malignant cells and in this respect, TfR is an attractive target for cancer therapeutics.
There are a number of engineered ligands that have been designed to specifically bind TfR and fall under the categories of small molecules, peptides, proteins or oligonucleotides. Lucidly, transferrin (Tf) has been extensively studied as a model ligand to observe the effect of protein corona on binding TfR, as well as engineered ligands, namely the peptides LT7 and DT7.
Notably, polystyrene nanoparticles (PNs) have served as effective therapeutic delivery vehicles and provided an adequate model of how polymeric nanoparticles behave in biological systems. Upon introduction to biological systems, ligand-modified PNs attract various blood proteins and subsequently form protein corona. In this respect, it is important to observe how ligand-modified PNs behave in the presence of protein corona which may negatively impact the binding abilities of the therapeutic-PN conjugation. With that said, there is disputed evidence that ligand-modified PNs suffer reduced efficacy upon introduction into the blood due to protein corona formation and the authors posture that the size and conformation of the ligand payload dictate corona composition. The authors also assert that corona composition will negatively dictate the efficiency and efficacy of the interaction between the vehicle and intended cell target.
In this publication, maleimide-functionalized PNs were bioconjugated with three known, thiolated TfR interactors; namely Tf, LT7 and DT7 as well as a negative control, PEG. Subsequently, these ligand-modified PNs (Tf-PN, LT7-PN, DT7-PN and PEG-PN) were incubated with 100% human plasma which was carried out to mimic the protein corona formed upon introduction into a biological system. Transmission Electron Microscopy, corona preparation, SDS-Page, MS and exocytosis assays were employed in tandem with OpenSPR experiments and the results from this study demonstrated that the formation of an in vitro protein corona masked the ability of the aforementioned ligands to effectively bind their target receptor, TfR.
The results obtained from surface plasmon resonance studies suggest that the targeting capacity of ligand-modified PNs were masked after the formation of in vitro protein corona.
Why was OpenSPR instrumental for this research?
Surface plasmon resonance (SPR) was used to understand the interactions between bioconjugated polystyrene nanoparticles and TfR. TfR was immobilized on a carboxyl sensor and the protein-nanoparticle experiment was run on an OpenSPR instrument. By using OpenSPR, the researchers were able to determine the increase or reduction in binding affinities between bare ligand-modified PNs, ligand-modified PNs incubated with human plasma and TfR. OpenSPR binding curves demonstrated that upon corona formation, all ligand-modified PN affinities reduced to the level of the negative control, PEG-PN. Importantly, this binding data suggests that protein corona formation may significantly reduce the interaction potential of ligand-modified PNs to target TfR, a common target for anti-cancer therapeutics. Thus, OpenSPR binding data provided insight into exploring more efficient drug delivery methods. By using OpenSPR, the researchers were able to get SPR data from their own bench, to help them accelerate their research and publish their discovery faster.
Why is SPR critical for publications? How does OpenSPR help?
SPR is a label-free technology which allows researchers to quantitatively analyze binding between two biomolecules. SPR technology allows us to determine the kon, koff and KD of interactions, providing deeper insight into binding events compared to other techniques that only give endpoint measurements, such as pull-down assays. SPR is necessary not only for publications but for the advancement of many fields of medicine and medical research as can be seen below with the significant increase in publications that rely on SPR data.
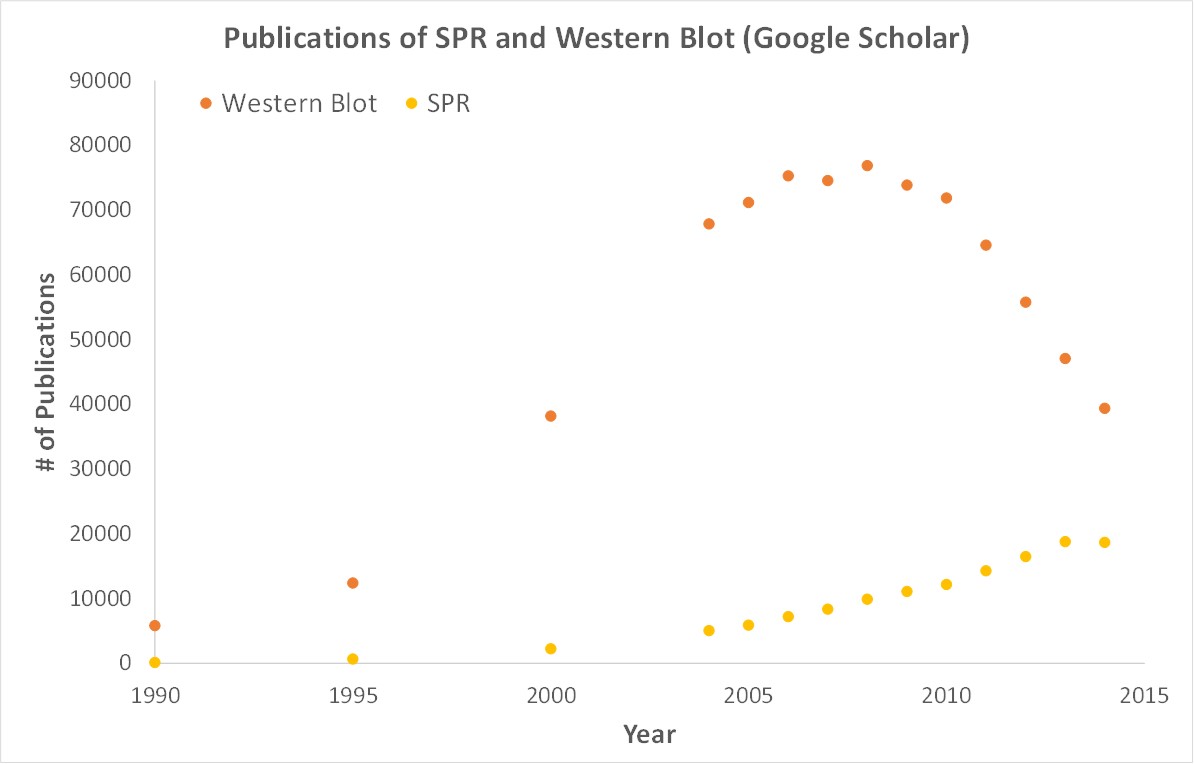
Scientific publications involving SPR have increased drastically over the years. SPR has become fundamental for publications while traditional techniques like Western Blots are becoming less important.
OpenSPR is a user-friendly and low maintenance benchtop SPR solution that is currently being used by hundreds of researchers. With access to SPR technology on your own lab bench you can get the high quality data you need to accelerate your research and publish faster.
