Glioblastoma (GMB) is a subtype of an Astrocytoma, known as the most aggressive form of brain cancer. Furthermore, Glioblastoma is a Grade IV tumour type that is characteristically the most malignant, fastest-growing, easily-metastasized, and efficient blood vessel producer out of all tumour types. These characteristics are some of the most important hallmarks of cancer. Since it is difficult to adequately treat patients who are suffering from glioblastoma it’s extremely necessary to develop adequate treatment to combat this tumour subtype. RNA binding proteins (RBPs) are an emerging source for glioblastoma targets. It is important to note that RNA binding protein, Musashi1 (Msi1), has been identified by multiple investigators as an extremely relevant oncogenic factor with respect to glioblastoma treatment.
In this new publication, Dr. Luiz Penalva et al. use the OpenSPR-XT’s automated, localized surface plasmon resonance technology to provide them with the key binding data needed for their latest discovery of an effective, small molecule glioblastoma treatment. The publication published in RNA Biology titled, “Luteolin inhibits Musashi1 binding to RNA and disrupts cancer phenotypes in glioblastoma cells” uses binding kinetics data generated from the OpenSPR-XT to assess the effectiveness of luteolin binding to RNA binding protein, Msi1.
About the Publication
Msi1 is one of over 1000 RNA binding proteins and maintains an important role in normal human gene expression. This 39 kDa RBP, which tends to be expressed in high levels in GMBs, constitutes an example of an “onco-RBP” due to the significant role it plays in abnormal cellular process including apoptosis, adhesion, and migration, among many others. It is important to note that onco-RBPs are gaining traction as highly attractive therapeutic targets to combat recurring GBMs because multiple oncogenic pathways can be disrupted at the same time. Moreover, luteolin is a 286 Da, low molecular weight phytochemical, commonly found in celery, green peppers, and chamomile, and retains anti-cancer properties. Luteolin has been shown to induce apoptosis in cancerous cells, preventing angiogenesis and progression of abnormal cellular cycles.
In this publication, the investigators began their project with high throughput screening (HTS) via biochemical fluorescence polarization. About 25,000 drug compounds were screened against Msi1 and this provided the investigators a leading candidate, luteolin. Subsequently, a cell viability assay was carried out with GBM cells, namely U251 and U343 cells, as they exhibit exceptionally high expression levels of Msi1. As a control, normal astrocytes were also selected for the cell viability assay to observe the potential cytocidal effects of luteolin on normal cells; notably, luteolin displayed no side effects on normal astrocytes but significantly reduced viability in U251 and U343 cells, perhaps indicating a high degree of selectivity for the RBP, Msi1.
To confirm suspected selectivity, U251 and U343 cells were subjected to Msi1 knockouts (KO). Upon administration of luteolin to the Msi1 KO cells, there were no significant changes in cell viability and in this respect, selectivity was confirmed. Following further confirmation of luteolin-Msi1 binding via NMR, and surface plasmon resonance was promptly recruited to further explore the kinetic parameters of this interaction.
Why was OpenSPR instrumental for this research?
Surface plasmon resonance (SPR) was used to quantitatively understand the interaction between Msi1 and luteolin. Untagged Msi1 was coupled to an carboxyl sensor chip via EDC/NHS coupling and 5 concentrations of luteolin were injected into the instrument via the OpenSPR-XT’s autosampler. Importantly, luteolin was demonstrated to bind Msi1 with an equilibrium dissociation constant (KD) of 3.2 μM and on/off rates of 6.16 X 103 M-1s1 and 1.97 X 10-2 s1, respectively. It is very important to note that OpenSPR-XT served as an automated and highly reproducible validation tool for characterizing the kinetic parameters of the interaction between the investigators leading candidate, luteolin, and onco-RBP Msi1. By using the OpenSPR-XT, the researchers were able to get SPR data from their own bench, to help them accelerate their research and publish their discovery faster.
Why is SPR critical for publications? How does OpenSPR help?
SPR is a label-free technology which allows researchers to quantitatively analyze binding between two biomolecules. SPR technology allows us to determine the kon, koff and KD of interactions, providing deeper insight into binding events compared to other techniques that only give endpoint measurements, such as pull-down assays. SPR is necessary not only for publications but for the advancement of many fields of medicine and medical research as can be seen below with the significant increase in publications that rely on SPR data.
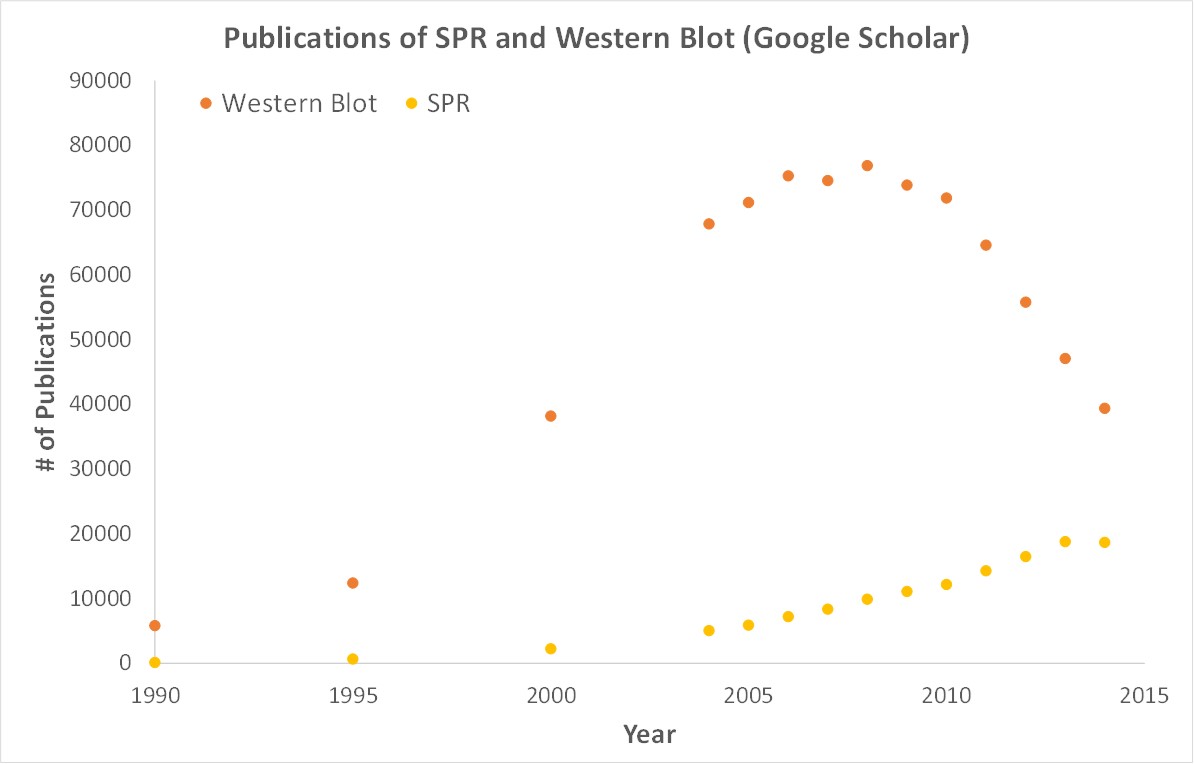
Scientific publications involving SPR have increased drastically over the years. SPR has become fundamental for publications while traditional techniques like Western Blots are becoming less important.
OpenSPR is a user-friendly and low maintenance benchtop SPR solution that is currently being used by hundreds of researchers. With access to SPR technology on your own lab bench you can get the high-quality data you need to accelerate your research and publish faster.
