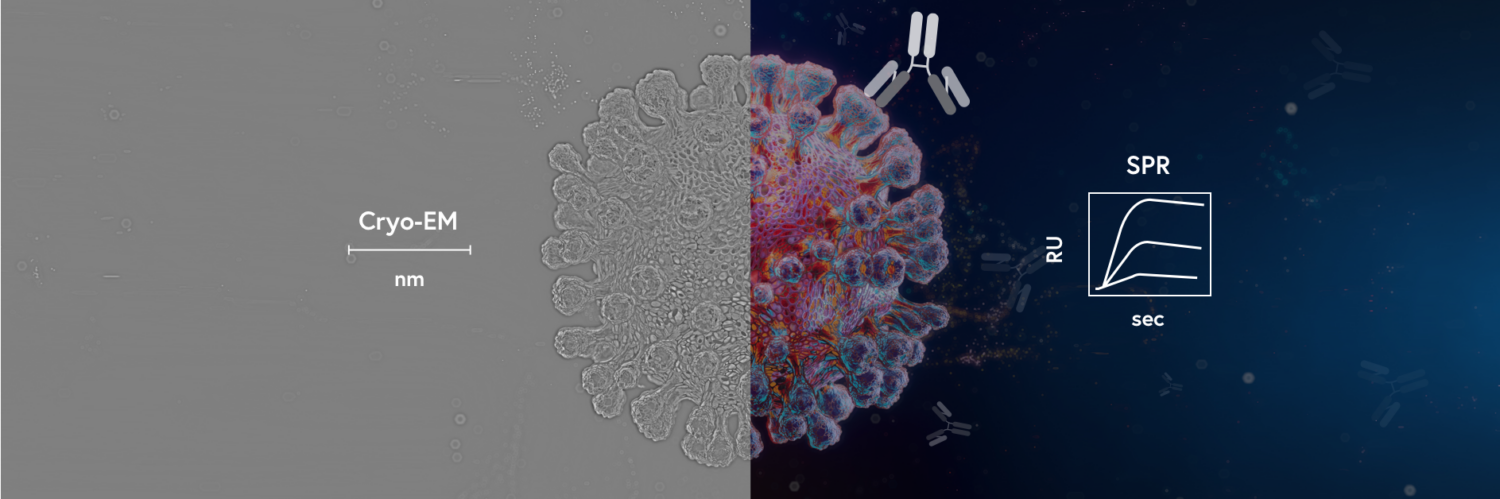
How did two cutting-edge technologies ultimately enable researchers to develop COVID-19 vaccines at an unprecedented rate?
The development of COVID-19 vaccines is a seminal discovery of this decade. The most remarkable feature of this achievement was the unprecedented rate at which the vaccines were developed. A major time-consuming bottleneck in vaccine development is the identification as well as robust structural and functional characterization of the target protein. With cryo-electron microscopy (cryo-EM) and surface plasmon resonance (SPR), scientists were able to rapidly characterize the structure and function of coronaviruses that ensured accelerated development of therapeutics and diagnostic solutions. In this blog post, you will find information on the advantages of using cryo-EM and SPR together to gain biomolecular structural and functional insights.
Blog Overview
- Overview: Cryo-EM and SPR technologies
- How cryo-EM and SPR technologies are accelerating coronavirus research?
- How are our SPR platforms unique?
- Resources to help you become an SPR expert
Overview: Cryo-EM and SPR technologies
Uncovering the structures of biomolecules is required to understand fundamental biological processes and to develop new medicines. Both X-ray crystallography and nuclear magnetic resonance spectroscopy have been the gold standard for understanding the structural identity of biomolecules. However, both methods require large sample sizes to process information, making them non-favorable options to map structures of scarcely available biomolecules. An additional disadvantage of X-ray crystallography is that it requires crystallization of biomolecules. Crystallization is a difficult process that makes the environment of biomolecules non-physiological. In contrast, cryo-electron microscopy (cryo-EM) — the technology that won the 2017 Nobel Prize in Chemistry — does not require large sample volumes or crystallization. This technology can capture images of flash-frozen biomolecules, revealing their 3D configuration in near-native states. Cryo-EM advantages have allowed scientists to map biomolecular configuration in near-atomic resolution, enabling them to understand how molecular machines work within our cells in ways that were previously impossible.
While understanding the structural identity of biomolecules is important, it doesn’t tell you the whole story of how biomolecules interact with each other. Biomolecular interaction analysis, in addition to structural information, is required for a comprehensive understanding of human development, disease pathogenesis, and to develop therapeutic interventions. There are different types of technologies that can measure biomolecular interactions, and thus it is imperative to choose the right technique for your experiments. Surface plasmon resonance (SPR) is a powerful technique that can analyze biomolecular interactions. It’s a label-free technique for measuring biomolecular interactions in real-time that is both flexible and information-rich as it provides detailed kinetic and affinity data on a myriad of biological molecules. As a highly sensitive technology, SPR can distinguish between different time-resolved binding profiles of key interactions for a wide range of applications.
These two technologies have been able to address the magnified need for novel therapeutics and diagnostic solutions to combat the current pandemic. The combined advantages of cryo-EM and SPR technologies have enabled scientists to rapidly characterize and identify coronavirus’ protein structure and binding partners that ultimately led to the development of different therapeutic solutions, including vaccines.
How cryo-EM and SPR technologies are accelerating coronavirus research
A hallmark research published in Science last year identified the structure and function of SARS-CoV-2 (Betacoronavirus) by using both cryo-EM and SPR1. Wrapp et al. used cryo-EM to decipher the structure of the SARS-CoV-2 spike protein (S) trimer in the prefusion conformation and showed that the predominant state of the trimer has one of the three receptor-binding domains (RBDs) rotated up in a receptor-accessible conformation. Since the S protein is required for the entry of SARS-CoV-2 into the host cells, understanding how the RBD domain of the S protein undergoes dynamic changes during this process can provide critical insights for the development of vaccines, therapeutic antibodies, and diagnostics. To further understand the binding partners of the S protein that can mediate viral infection, the authors then used SPR to analyze biophysical interactions and showed that the S protein can bind angiotensin-converting enzyme 2 (ACE2). The authors also showed that the SARS-CoV-2 S protein binds ACE2 with higher affinity than SARS-CoV-1 S protein, which could contribute to the apparent ease with which SARS-CoV-2 can spread from human to human.
Another publication by Song et al. in Nature Communications highlighted how these two complementary technologies can further advance coronavirus research2. As mentioned earlier, it is now well-known that the S protein of coronaviruses mediates viral entry into host cells by binding to host receptors. However, how the RBD of the S protein undergoes conformational changes for receptor recognition has not been elucidated in Alphacoronavirus. The authors used cryo-EM to reveal the structure of the HCoV-229E S trimer in the perfusion state, and provided insights into how closed versus open conformation of the S glycoprotein can determine receptor binding activity in Alphacoronavirus. The authors then used our SPR platforms to show that the conformational transformation from closed to activated state can lead to differential interactions of the RBD of the S protein.
These publications highlight the advantages of using cryo-EM and SPR together to generate structural and functional insights into the molecular mechanisms of how biomolecules work.
How are our SPR platforms uniquely suited to accelerate your R&D?
SPR is a gold standard technique for analyzing biomolecular interactions because of the high-quality binding affinity and kinetics data this technology can enable you to generate. However, SPR has remained fairly inaccessible as a result of operational complexity and cost. To disrupt this status-quo, we leveraged nanotechnology and digital microfluidics to develop innovative and accessible SPR platforms for rapid, reliable, and label-free characterization of biomolecules.
We first designed the OpenSPR platform that has the same benefits of traditional SPR but is more affordable, fits on your laboratory benchtop, requires lower maintenance, has better protection against vibration and noise, and less interference from buffer mismatch and temperature drift. For the first time, scientists were able to use SPR technology right at their benchtop!
In our continued efforts to support and help scientists succeed, and to create bigger impacts, we then designed Alto – the world’s first digital SPR platform. Alto can accelerate all stages of your research programs by providing you with a fully-automated analysis of key biomolecular interactions. By harnessing the superior liquid handling abilities of digital microfluidics, Alto provides you with high-quality, label-free kinetic analysis using only 2 µl sample volume. Our SPR platforms are currently empowering over 600 scientists from global biotech, pharmaceutical and research organizations to push the limits of their research and bring novel healthcare solutions to market.
Resources to help you become an SPR expert
- Learn more about the technologies (nanotechnology and digital microfluidics) used in our SPR platforms.
- What are the best practices of SPR experiments?
- Guide: how to select the best SPR sensor.
References
- Wrapp D, Wang N, Corbett KS, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260-1263.
- Song X, Shi Y, Ding W, et al. Cryo-EM analysis of the HCoV-229E spike glycoprotein reveals dynamic prefusion conformational changes. Nat Commun. 2021;12(1):141.