Our next generation SPR platforms provide high-quality data and streamline research workflows to give you an edge in the fight against influenza.
Influenza viruses cause respiratory infections in humans and are responsible for annual epidemics and historical pandemics, presenting significant health, societal, and economic burdens.1 As their RNA genome lacks proofreading capabilities and continually accumulates mutations, influenza viruses are always evolving.2 As such, effective monitoring of this viral evolution is essential for preparing for and mitigating potential outbreaks.
A network of scientists, part of the Global Influenza Surveillance and Response System (GISRS) coordinated by WHO, continually monitor influenza subtypes and provide recommendations for vaccine formulation updates by characterizing influenza virus strains and predicting potential dominating strains.2 While we have made strides in virus surveillance, vaccine development and treatments, newer challenges are emerging. Increasing population, human mobility, and mounting environmental pressures all increase the risk of zoonotic infections and rates of transmission. Tools that provide accurate and comprehensive data while streamlining research workflows provide important opportunities for improving viral research.
In this blog, we discuss how Nicoya’s surface plasmon resonance (SPR) platforms, OpenSPR™ and Alto™, can be applied in the discovery and characterization of influenza-related targets to keep up with viral evolution.
Table of contents
- Influenza A viral antigens
- Keeping up with antigenic drift
- Characterization with surface plasmon resonance (SPR)
- Nicoya’s next-generation localized SPR (LSPR)
- Conclusion
Influenza A viral antigens
One of the most prominent viral pathogens affecting the human populace is influenza, or the “flu” with types A and B responsible for seasonal flu epidemics.3 Influenza A is the only known type to have caused flu pandemics, having instigated all five historical occurrences since 1900.3
The interplay between the influenza virus and the host immune response results in a phenomenon known as antigenic drift, or mutations at antigenic sites, that may permit the virus to evade detection from immune control. Consequently, the influenza virus is continually evolving.2 This fitness advantage provides the virus the chance to emerge as the novel dominant strain, replacing the circulating strain for which the populace may already be vaccinated.2
Of the influenza virus anatomy, two of the most important components to characterize are the Influenza A Nucleoprotein (NP) and Influenza A Hemagglutinin (HA), that contribute to infection and replication.
Influenza A nucleoprotein (NP)
Influenza A viral nucleoprotein (NP) is a structural protein that plays a key role in viral replication and host adaptation. NP is highly conserved among influenza strains and is implicated in myriad functions, making it an ideal target for diagnostic testing and therapeutic intervention. Antibodies targeting NP proteins are commonly used for immunodetection of influenza viruses in various assays, including enzyme linked immunosorbent assays (ELISA), lateral flow assays (LFA), and direct fluorescent antibody tests. Due to the high frequency of antigenic drift or shift among different influenza strains, broad-spectrum influenza antibodies are particularly desired for flu diagnostics.
Influenza A hemagglutinin (HA)
Influenza A hemagglutinin (HA) is a membrane glycoprotein implicated in viral infection. As it functions by binding to the target cell receptor and triggering endocytosis, its receptor-binding domain (RBD) is critical for vaccine development and a popular target for antibody therapeutics. Monitoring mutations of this protein is essential for continual development of vaccine formulations and predicting when a strain may become human-adapted and pose a pandemic risk.4
Keeping up with antigenic drift
Effective viral surveillance is imperative to keeping up with antigenic drift, mitigating annual epidemics, and preparing for future outbreaks. However, several challenges exist in keeping up with antigenic drift. Recommendations on vaccine strains must be made 6-8 months in advance of producing vaccines and subsequently inoculating the populace. During this time, the dominant strain may change as a result of continued antigenic drift.2 The emergence of challenges such as increasing human population and mobility, and mounting environmental pressures, have also added to the burden of viral surveillance by increasing the probability of virus transmission from zoonotic reservoirs and increasing the spread of influenza.5-7
To monitor antigenic drift and stay ahead of novel strains before they emerge as the dominant virus, scientists often focus on characterizing key proteins associated with virulence to better understand how they function and how to target them. The data obtained are imperative for devising annual vaccine formulation updates and novel therapeutics.
Traditional techniques such as ELISA and western blot (WB) are time-consuming, depend on the use of tags for analysis, provide limited end-point data, and require large sample volumes. Later methods such as glycan microarrays developed to evaluate the glycan–HA specificity require high virus levels and don’t provide kinetic parameters.8 To better characterize antigenic drift and stay ahead of the virus, there is a need for more efficient tools that provide comprehensive data and simplify research workflows.
Novel label-free biosensing technologies have emerged over the past few decades, of which SPR-based analyses have garnered considerable attention for characterizing biomolecular interactions.4
Characterization with surface plasmon resonance (SPR)
An alternative method that has gained traction in the past few years for characterization of influenza targets is SPR, a streamlined, label-free and real-time technique that provides comprehensive affinity and kinetics data. Previous work by Suenaga and colleagues on HA-glycan binding conducted on a conventional SPR platform demonstrated the method’s suitability for viral surveillance by monitoring HA mutations.4,9 In addition to being highly sensitive and providing accurate affinity data, SPR also provides kinetics data, association rates (ka), dissociation rates (kd) and equilibrium dissociation constant (KD), providing a more accurate picture of the interaction between viral antigens and their targets. However, it’s important to note most SPR platforms on the market have high operational costs and require extensive training and years of technical experience to produce high-quality data.
Nicoya’s next-generation localized SPR (LSPR)
Our benchtop instruments leverage localized SPR (LSPR) technology for a more powerful and efficient alternative. Designed with the user in mind, they shorten the learning curve of using SPR by automating tedious tasks and eliminating the guesswork of assay design and data analysis with intuitive software. To demonstrate the capabilities of our SPR platforms for influenza-related applications, we teamed up with Sino Biological to conduct kinetic analysis and epitope binning studies using their recombinant products.
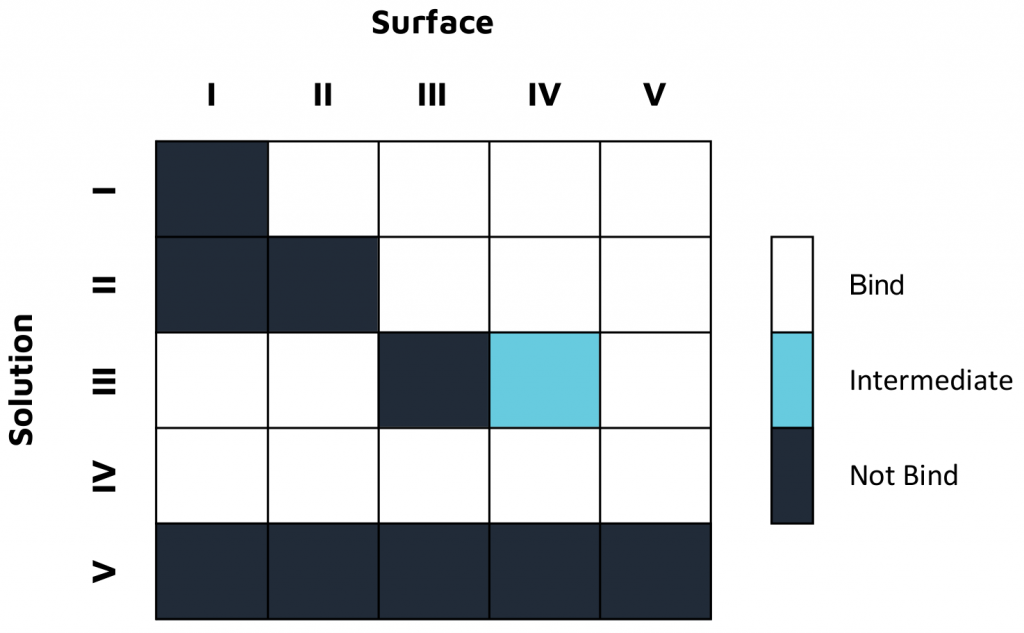
In one study, characterization experiments were conducted with a 16-channel digital SPR system to analyze five unique antibodies against two influenza A NP proteins, and two unique antibodies against an influenza A HA protein, with the results summarized in Figure 1. Alto, the first SPR platform to integrate digital microfluidics (DMF) technology, was proven to streamline kinetics analysis and epitope binning of the antibodies of interest by providing automated on-cartridge serial dilutions and requiring only 2 µL of sample. Analysis of the antibodies was completed within one week, with each experiment requiring less than 20 minutes of hands-on time. Read the full application note here.
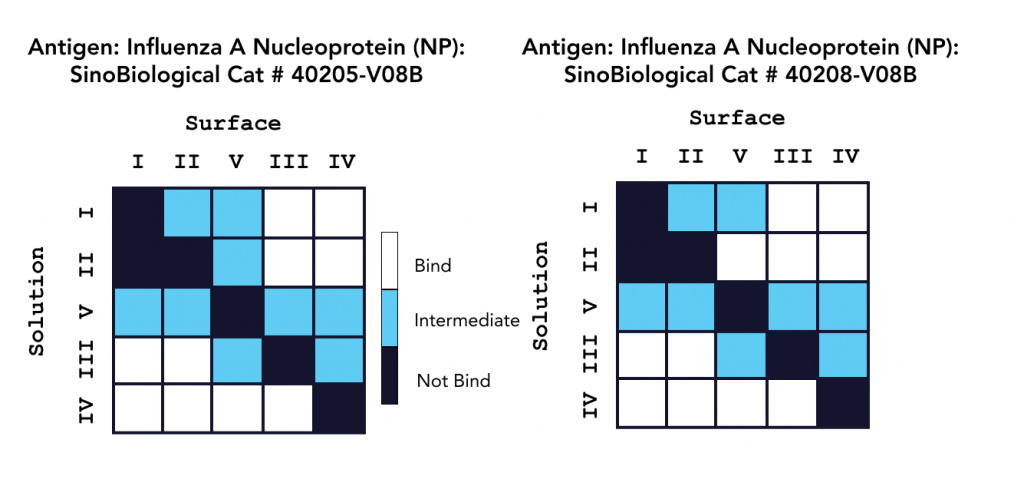
In a second study, epitope characterization studies were conducted on OpenSPR in a classical binning format with five unique antibodies against an influenza A NP protein, with the results summarized in Figure 2. This 2-channel system also successfully provided the necessary kinetic data to successfully understand, rank, and optimize these antibodies for therapeutics development, without the use of tags or labels. Read the full application note here.
Both platforms successfully characterized Sino Biological’s influenza antibodies, highlighting the applicability and efficiency of LSPR for epitope characterization. The comprehensive data generated in both studies provide a deeper understanding of both antibody efficacy and antigen drift for the development and optimization of vaccines and antibody therapeutics.
Conclusion
Continued surveillance of antigenic drift is imperative to keeping up with viral evolution and designing effective vaccines and therapeutics. Tools that provide accurate and comprehensive data while streamlining research workflows provide important improvements in surveillance.
OpenSPR and Alto successfully characterized Sino Biological’s highly specific influenza antibodies through epitope binning and kinetic determination. Compared to traditional methods of characterization, our SPR platforms accelerate the analysis of multiple interactions by eliminating the need for labels and reducing time-consuming preparation steps, while providing comprehensive affinity and kinetics data. Alto’s ultra-low sample consumption, automated on-cartridge serial dilutions, and high-throughput capabilities further streamline the experimental workflow.
Our SPR platforms are well-aligned to meet the needs of modern researchers looking to keep up with viral evolution and develop new therapeutic strategies without compromising on data quality. Learn how you can level up your antiviral research by chatting with one of our application scientists today!
Viruses evolve, and so should your lab. Switch to SPR today.
References
- Becker T, Elbahesh H, Reperant LA, Rimmelzwaan GF, Osterhaus ADME. Influenza vaccines: Successes and continuing challenges. J Infect Dis. 2021;224(12 Suppl 2):S405-S419. doi: 10.1093/infdis/jiab269
- Webster RG, Govorkova EA. Continuing challenges in influenza. Ann N Y Acad Sci. 2014;1323(1):115-139. doi:10.1111/nyas.12462
- Types of Influenza Viruses. Centers for Disease Control and Prevention. Updated December 2, 2022. Accessed January 13, 2023. https://www.cdc.gov/flu/about/viruses/types.htm
- Suenaga E, Mizuno H, Kumar PKR. Influenza virus surveillance using surface plasmon resonance. Virulence. 2012;3(5):464-470. doi: 10.4161/viru.21822
- Findlater A, Bogoch II. Human mobility and the global spread of infectious diseases: A focus on air travel. Trends Parasitol. Vol. 2018;34(9):772-783. doi: 10.1016/j.pt.2018.07.004.
- Marani M, Katul GG, Pan WK, Parolari AJ. Intensity and frequency of extreme novel epidemics. PNAS. 2021;118(35):e2105482118. doi: 10.1073/pnas.2105482118.
- Mora C, McKenzie T, Gaw IM, et al. Over half of known human pathogenic diseases can be aggravated by climate change. Nat Clim Chang. 2022;12(9):869-875. doi: 10.1038/s41558-022-01426-1.
- Childs RA, Palma AS, Wharton S, et al. Receptor-binding specificity of pandemic influenza A (H1N1) 2009 virus determined by carbohydrate microarray. Nat Biotechnol. 2009;27(9):797–799. doi: 10.1038/nbt0909-797
- Suenaga E, Mizuno H, Penmetcha KKR. Monitoring influenza hemagglutinin and glycan interactions using surface plasmon resonance. Biosens Bioelectron. 2012;32(1):195-201. doi: 10.1016/j.bios.2011.12.003