Read on to learn about the two most common approaches for measuring binding kinetics, multi-cycle kinetics (MCK) and single-cycle kinetics (SCK), and how to select the right one for your interaction.
Introduction
The study of biomolecular interactions is critical to our understanding of biological systems and the forces involved in driving biological action. Biomolecular interaction analysis (BIA) enables us to examine how specific biomolecules interact, providing us with important information about their mechanism and behaviour in living systems. With the versatility to be applied on many biomolecular pair types, including antigens and antibodies, drugs and proteins, and hormones and receptors, BIA has been an accelerator of many discoveries in medical, pharmaceutical and fundamental biological research.
Among the parameters that can be measured, kinetic analysis is a powerful method for elucidating the rate at which an interaction occurs. Observing how quickly a biomolecular complex forms and comes apart, or the association and dissociation rates of the interaction, can give insights about the roles of different biomolecules, residence times for antibody and receptor-target complexes, competitor drugs’ mechanisms, and much more1. To learn more about the importance of binding kinetics, check out our blog on What Binding Kinetics Can Tell You That Affinity Can’t.
Tools such as surface plasmon resonance (SPR) have advanced our ability to perform kinetic studies by measuring interactions in real-time and in a label-free environment. In an SPR experiment, there are two common experimental methods for kinetic analysis: Multi-cycle kinetics and single-cycle kinetics. Before selecting a format for your next experiment, it is important to understand how they differ and which is best for your specific interaction.
Blog Overview
- What is multi-cycle kinetics?
- What is single-cycle kinetics?
- Which method is best for my interaction?
- Analyze binding kinetics with Alto
- MSK and SSK analysis of a protein-antibody interaction on Alto
What is multi-cycle kinetics?
To perform an SPR experiment, one of the interactants, referred to as the ligand, is immobilized on a sensor surface, and the second interactant, referred to as the analyte, is passed over the surface via a fluidic system. The response change over time is plotted, and the resulting sensorgrams are analyzed using analysis software equipped with various fitting algorithms. Experiments are performed in replicates, with 3-5 concentrations of each analyte being studied. A typical sensorgram includes a baseline phase where bulk flow (buffer) response is recorded, an association phase during which analyte is passed over the immobilized ligand, a dissociation phase where buffer flow resumes and bound analyte is removed from the surface according to its off-rate, and in cases where dissociation is too slow to go to completion on a reasonable experimental time scale, a regeneration step.
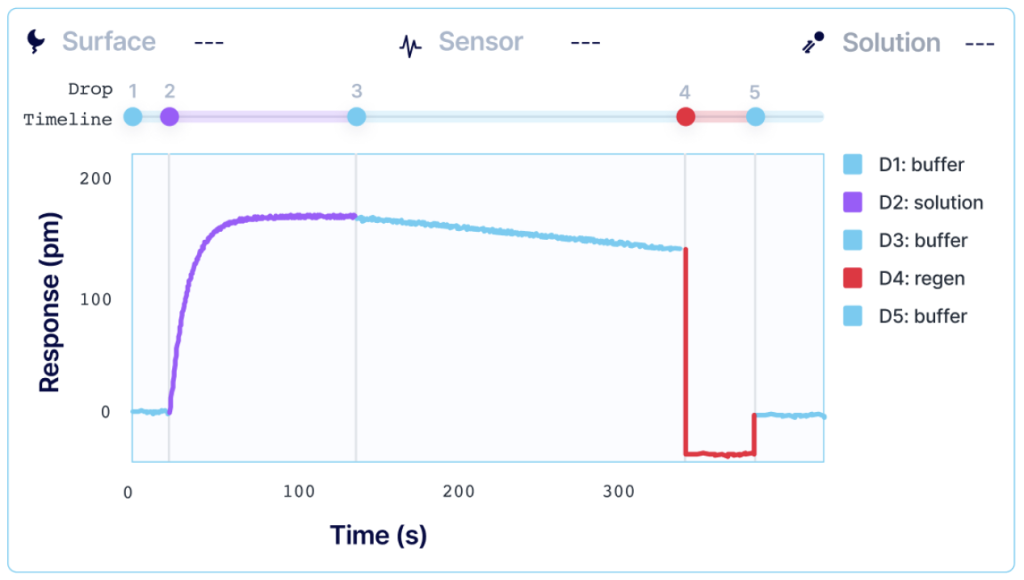
Kinetic analysis that is performed with alternating cycles of analyte injections and surface regeneration is known as multi-cycle kinetics (MCK). MCK is the most common strategy for determining interaction kinetics. Each injection of an analyte is done in a separate sequence, and a single SPR curve is generated per analyte concentration. High-throughput systems, such as Alto, allow users to automate MCK assays when working with a larger library of samples.
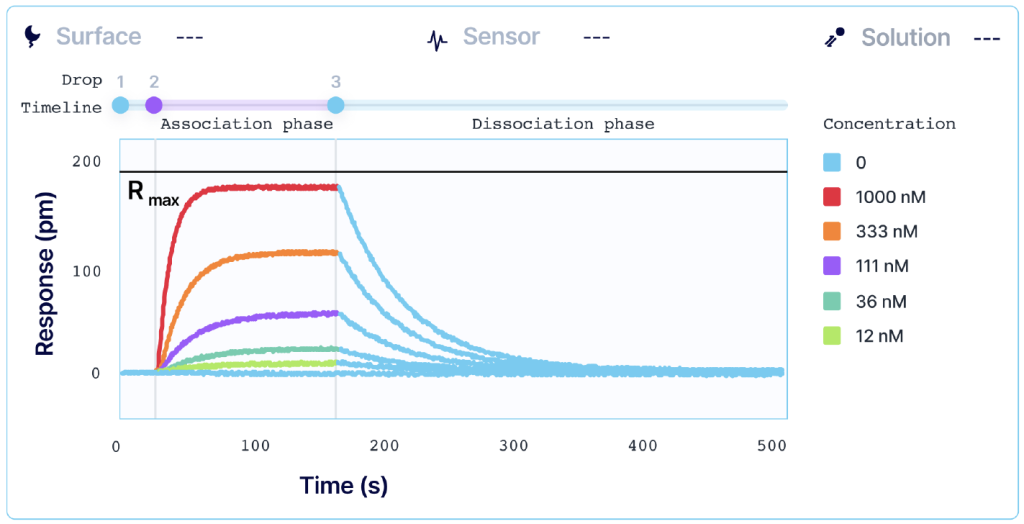
However, regeneration of your surface requires conditions that can sometimes damage the functionality of your ligand or irreversibly inactivate it. For systems where total regeneration is difficult to achieve, single-sequence kinetics may be a more optimal approach.
What is single-cycle kinetics?
Kinetic titration, also known as single-cycle kinetics (SCK), consists of sequential injections of increasing concentrations of the analyte over the ligand, without dissociation or regeneration between each sample concentration. The last (highest) analyte concentration is then followed by a single, long dissociation phase.
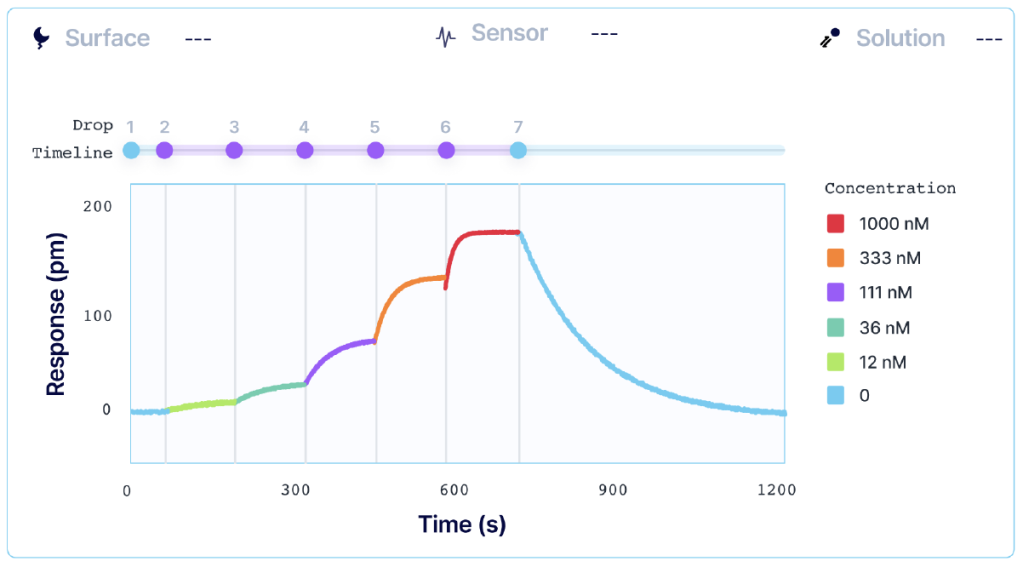
SCK reduces the analysis time for a given interaction, with the exact time depending on the relative length of the different steps (baseline, association, etc.) of the interaction, and whether a ligand capture approach is used. In cases where the total number of ‘samples’ (including actual analyte and regeneration solutions) is limited, the throughput budget can be increased by removing the recurring regeneration steps, and if applicable, ligand recapture of MCK.
In addition to a shortened assay run time, another advantage of the SCK method is that it requires fewer regeneration steps. This allows for the characterization of binding to ligand surfaces that are difficult to regenerate or where regeneration may inactivate the ligand, thus reducing the risk of damaging the ligand when using the sensor for multiple analyte concentrations2. SCK is also useful for capture methods as it removes the need to recapture the ligand between analyte concentrations.
Which method is best for my interaction?
One major disadvantage of SCK, especially for interactions with complex binding kinetics, is the reduced informational content obtained from the single dissociation phase of a given interaction. If there are concerns with the fits obtained from an SCK experiment, an MCK experiment may still be required for an easier diagnosis and better understanding of the underlying phenomena, due to the availability of multiple binding curves. For example, in an MCK experiment, a buffer blank is typically performed and subtracted from each of the individual analyte binding curves to correct for a drift in the baseline. As well, if a poor analyte injection occurs, the user can omit the data from the curve overlay during fitting. Thus, both MCK and SCK methods have their advantages and disadvantages, and one may be preferable over the other for a given application.
Analyze binding kinetics with Alto
Alto is a high-throughput benchtop SPR instrument that is powered by digital microfluidics (DMF), a liquid handling technology capable of accurately controlling and manipulating discrete nanoliter droplets with applied voltage. The versatility and automation provided by DMF gives users wide flexibility over assay design and makes Alto a powerful tool for generating high precision binding kinetics.
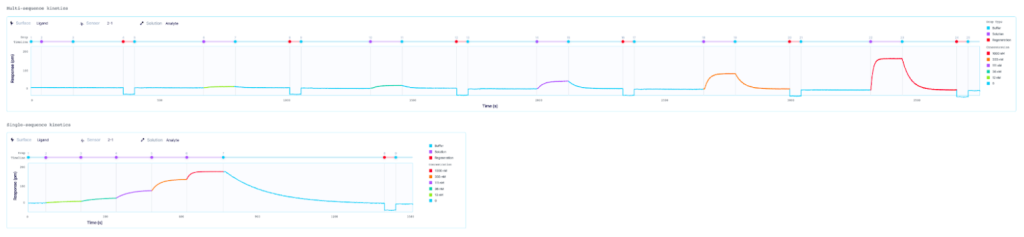
The analysis of biomolecular interactions on Alto differs from traditional SPR platforms in that it operates with sequences of fluidic droplets rather than fluidic cycles. ‘Sequence’ refers to the order and type of droplet that reaches the sensor throughout the experiment. As such, Alto’s multi-sequence kinetics (MSK) and single-sequence kinetics (SSK) methods offer the same functionality as traditional MCK and SCK assays, with added flexibility and control over assay design.
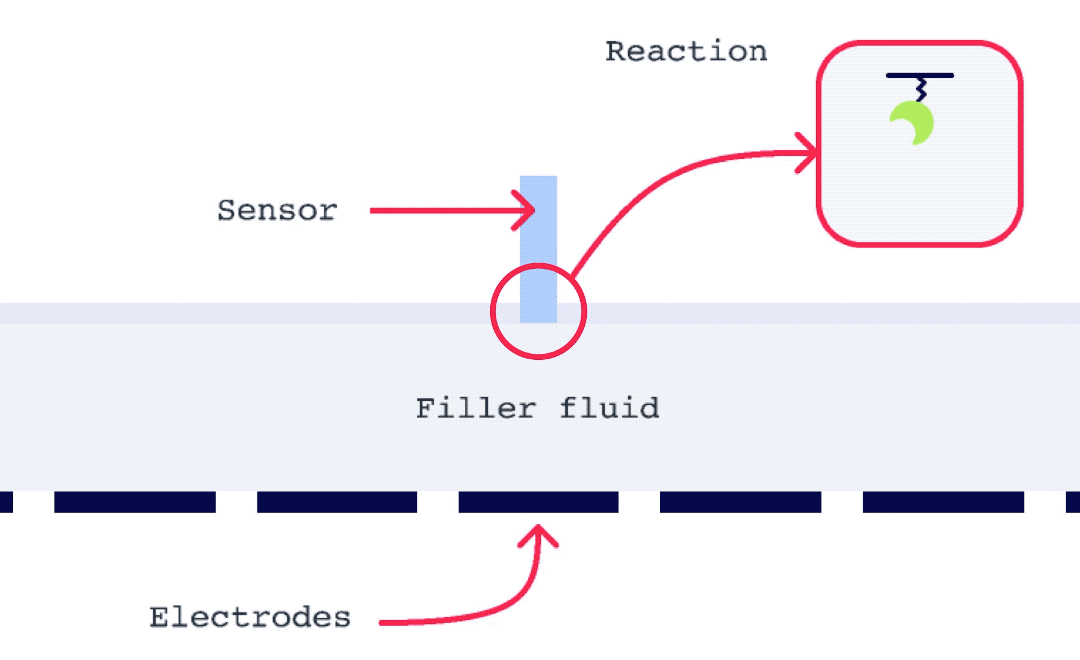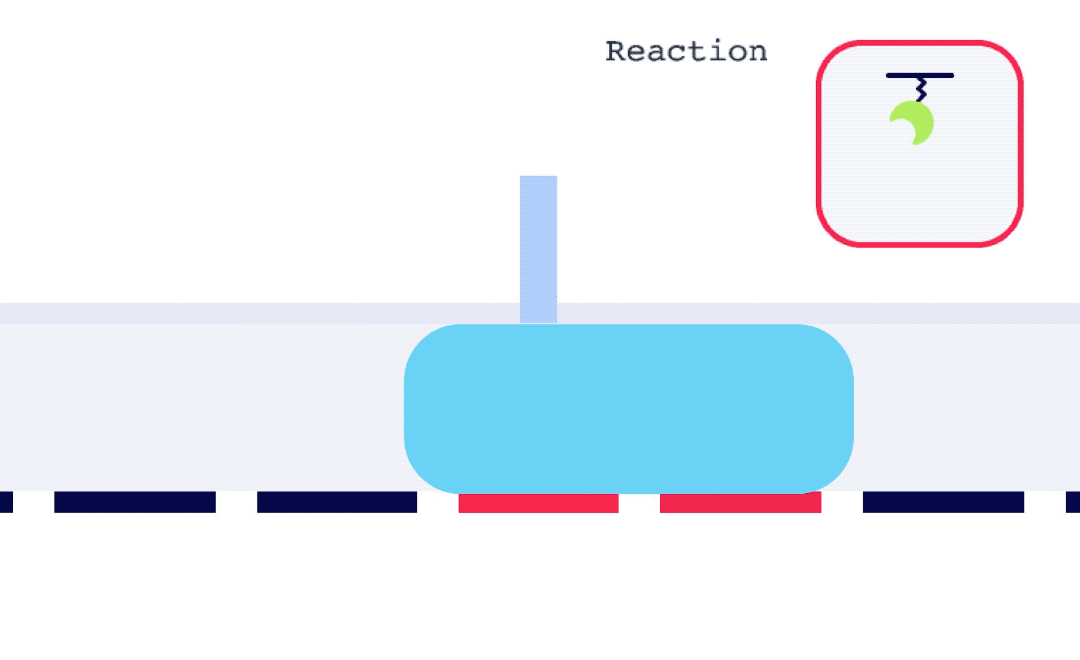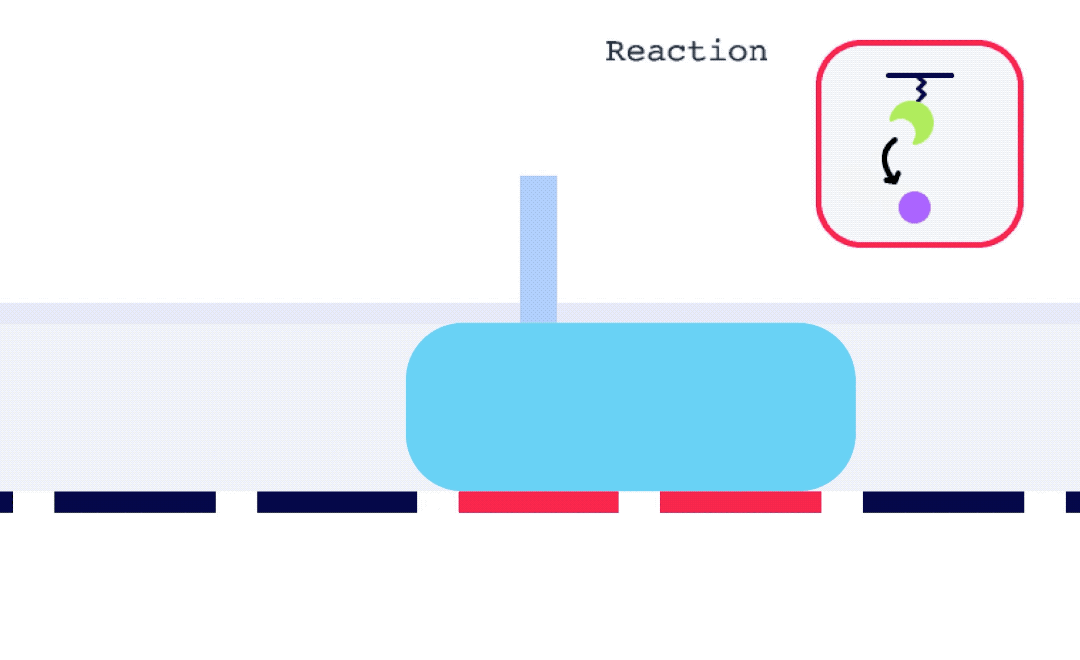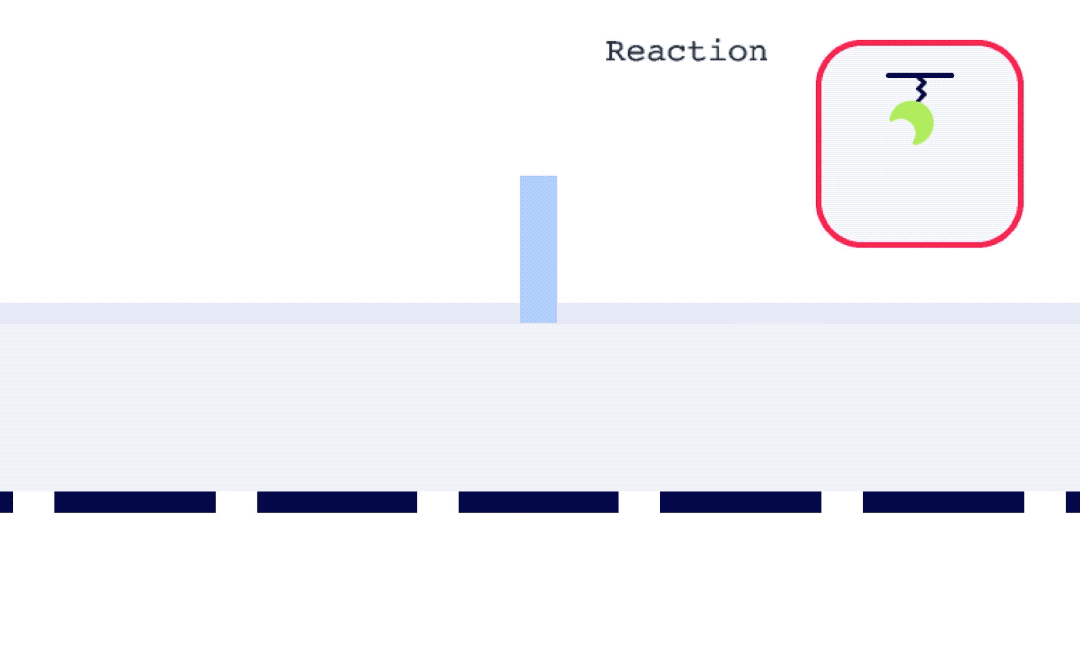
Alto is uniquely suited for SSK analysis as it enables the exchange of droplets on the sensor surface without the need for buffer flow between samples. As well, with each droplet being individually addressed within Alto’s ecosystem, the user has greater flexibility with their data. This allows for any risk from a compromised portion of an SSK experiment to be mitigated by simply removing that droplet segment from the analysis. Alto’s automated serial dilutions and digital microfluidic handling also make the instrument less prone to user errors and data artifacts caused by air or contamination, as seen in traditional fluidic systems.
MSK and SSK analysis of a protein-antibody interaction on Alto
Our recent application note demonstrates Alto’s ability to analyze samples and measure binding kinetics using these two different assay methods. Binding kinetics of Protein A with Human Immunoglobulin G (HIgG) were measured on Alto with both single-sequence and multi-sequence kinetics. The kinetic binding constants obtained with the SSK method were consistent with those measured with the MSK assay for the same interaction and analyte concentrations of Protein A binding with Human Immunoglobulin G (HIgG). Read the full application note.
Conclusion
Selecting the best assay format for your interactions will allow you to expand and optimize your kinetic studies and enable you to dig deeper into the roles and behaviours of a wide range of biomolecular targets. Alto’s versatile fluid handling makes it a powerful tool for a variety of SPR assay designs and applications to generate high precision binding kinetics, including novel and complex workflows not currently (or physically) available in traditional fluidic systems.
To learn more about Alto, download our brochure. Or, get started with kinetic experiments today by contacting our application scientists.
Get started with kinetic experiments today.
CONTACT APPLICATION SCIENTISTReferences
- Zheng, X., Bi, C., Li, Z., Podariu, M., Hage, D. S. (2015). Analytical methods for kinetic studies of biological interactions: A review. Journal of Pharmaceutical and Biomedical Analysis, 113, 163-180. https://doi.org/10.1016/j.jpba.2015.01.042
- Karlsson, R., Katsamba, P. S., Nordin, H., Pol, E., & Myszka, D. G. (2006). Analyzing a kinetic titration series using affinity biosensors. Analytical biochemistry, 349(1), 136–147. https://doi.org/10.1016/j.ab.2005.09.034
