In both industrial and clinical settings, biofilm formation by opportunistic human pathogens represents a significant area of concern. Biofilms tend to be composed of extracellular polymeric substances including proteins, DNA, and polysaccharides. They provide a strong layer of defense against antibiotics, immune factors, and physical environmental threats. Notably, these 3-dimensional polymeric films can be found on medical devices such as catheters, cardiac implants, and inadequately sterilized surgical tools and often cause hospital-acquired (nosocomial) infections.
Importantly, Psuedomonas aeruginosa is an opportunistic human pathogen which is culpable for a large number of nosocomial and chronic lung infections observed in patients. P. aeruginosa possesses an impressive arsenal of virulence factors, antimicrobial resistance mechanisms, and has the ability to form robust biofilms in a multitude of environments. Biofilm production and maintenance in P. aeruginosa is largely due to the highly complex and adaptable genetic regulation network, so it is important to understand the genetic regulation of biofilm production and maintenance.
In this new publication, Dr. Tung T. Hoang et al. used OpenSPR’s localized surface plasmon resonance technology to provide them with the key binding data needed for their latest discovery on biofilm formation in P. aeruginosa. The publication titled, “Novel dual regulators of Pseudomonas aeruginosa essential for productive biofilms and virulence” uses binding kinetics data generated from the OpenSPR to quantify the formation of a stable complex between 2 co-regulators, PA1226 and PA1413, and their target DNA binding sequence.
About the Publication
The complex network of gene regulation for biofilm production in the clinically relevant bacteria species, P. aeruginosa, is not well understood. Since this organism can form adaptive and persistent biofilms on biomedical devices and causes life-threatening lung infections in the elderly and immunocompromised individuals, it is important to understand the complex factors that contribute to genetic regulation. Probable transcriptional regulators, PA1226 and PA1413, are two DNA-binding proteins found in P. aeruginosa. As their names suggest, these proteins are responsible for regulating transcription of genes associated with biofilm production and maintenance. Strikingly, both of these proteins have been individually implicated in genetic regulation of biofilm production, even though there has been little to no evidence that suggests they act cooperatively.
In this publication, the investigators used transcriptomic technology to screen and identify hundreds of possible transcriptional regulators responsible for biofilm production from 42 P. aeruginosa mutant strains. Upon completion of the screening and subsequent crystal violet assays, an insertional mutation in the gene encoding PA1226 caused the most significant reduction in biofilm formation. Subsequently, D. melanogaster and BALB/c mouse infection models were used for in vivo studies and the results of the crystal violet assays were confirmed; P. aeruginosa PA1226 mutants were unable to effectively colonize their hosts.
Upon ChIP-seq and microarray analysis, electrophoretic mobility shift assays were carried out. Upon receipt of the results which were quite surprising, the investigators suggested that there must be a co-regulator necessary for the interaction between PA1226 and DNA to occur. It is important to note that this was a significant event in the study. Following co-immunoprecipitations and Ion-Trap LC-MS, it was strongly presumed that PA1226 and PA1413 act cooperatively to form an exceptionally stable regulator-coregulator-DNA complex. Afterwards, surface plasmon resonance was used to further explore this discovery.
Why was OpenSPR instrumental for this research?
Surface plasmon resonance (SPR) was used to quantitatively understand the co-regulating interaction between PA1226, PA1413, and DNA. His-tagged PA1413 was coupled to an NTA sensor chip and 5 concentrations of untagged PA1226 and DNA were independently injected into the instrument as negative controls. Subsequently, 5 concentrations of a mixture of PA1226 and DNA were injected into the instrument to test the hypothesis that a cooperative interaction is necessary for the transcriptional regulators to effectively bind DNA. Importantly, PA1226 and DNA showed no interaction with PA1413 when run independently, but the mixture of PA1226-DNA yielded highly concentration-dependent binding data and provided an equilibrium constant of 0.31 μM. The 3 lowest concentrations (33, 100, and 200 nM) of the PA1226-DNA fully dissociated from the sensor but the 2 highest concentrations (300 and 600 nM) of PA1226-DNA did not dissociate from the sensor. These findings corroborated binding data obtained from CoIPs and electrophoretic mobility shift assays.
In this respect, OpenSPR binding data validated the discovery that a regulator-coregulator-DNA complex forms between PA1226, PA1413, and DNA required for biofilm production in P. aeruginosa. By using OpenSPR, the researchers were able to get SPR data from their own bench, to help them accelerate their research and publish their discovery faster.
Why is SPR critical for publications? How does OpenSPR help?
SPR is a label-free technology which allows researchers to quantitatively analyze binding between two biomolecules. SPR technology allows us to determine the kon, koff and KD of interactions, providing deeper insight into binding events compared to other techniques that only give endpoint measurements, such as pull-down assays. SPR is necessary not only for publications but for the advancement of many fields of medicine and medical research as can be seen below with the significant increase in publications that rely on SPR data.
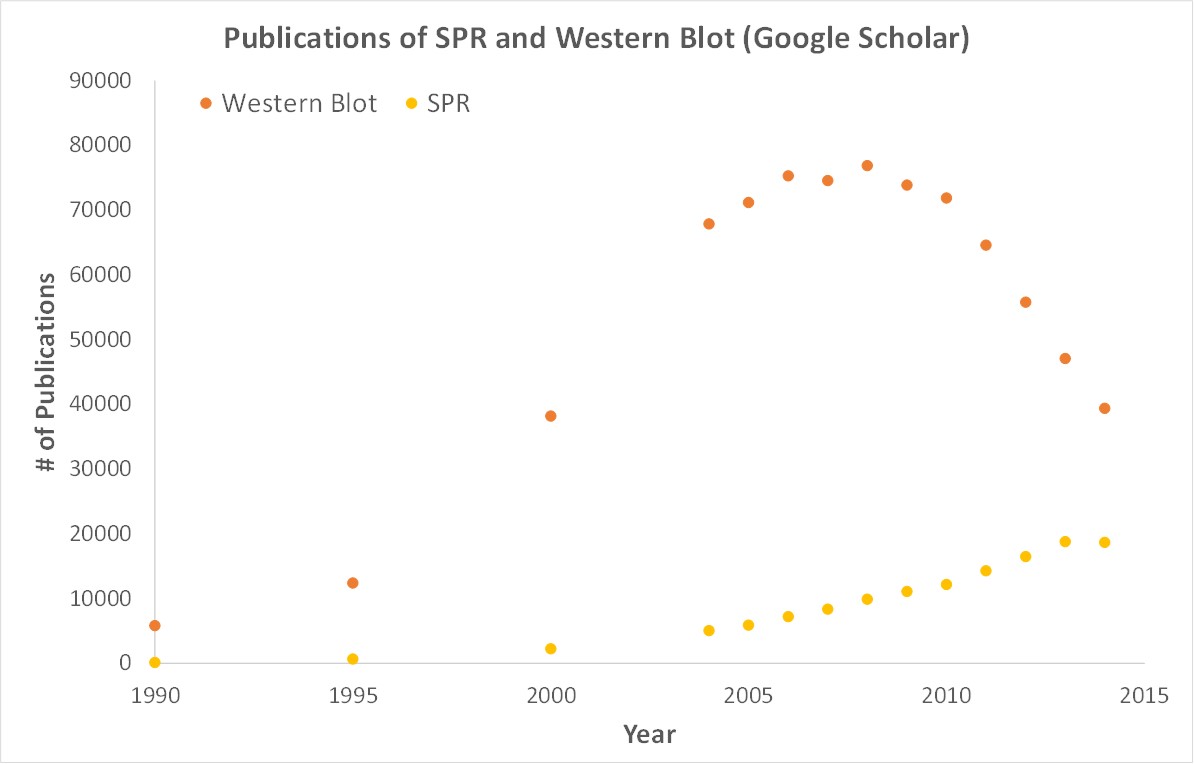
Scientific publications involving SPR have increased drastically over the years. SPR has become fundamental for publications while traditional techniques like Western Blots are becoming less important.
OpenSPR is a user-friendly and low maintenance benchtop SPR solution that is currently being used by hundreds of researchers. With access to SPR technology on your own lab bench, you can get the high-quality data you need to accelerate your research and publish faster.
