High-affinity SARS-CoV-2 antibody characterized using Surface Plasmon Resonance (SPR).
Finding a Solution to COVID-19 Together
Widely considered the largest public-health emergency of the last century, COVID-19 has affected billions of lives worldwide. The specific virus causing the current global crisis is SARS-CoV-2, a novel strain of a family called coronaviruses that often attack the respiratory system. The development of a vaccine to prevent the spread of SARS-CoV-2 is a priority for researchers worldwide.
As the scientific community steps in to end the crisis, it is crucial that researchers have access to the tools and technology that they need to combat the virus. Surface Plasmon Resonance (SPR) has long been considered one of the gold-standard techniques for key measurements involved in virology research by providing detailed binding data. This binding characterization is used in basic research for understanding viral infection mechanisms and is important in the development of therapeutics, vaccines, and the realization of effective diagnostic tools.
Kinetic analysis of a SARS-CoV-2 Antibody with the Spike Protein Receptor Binding Domain
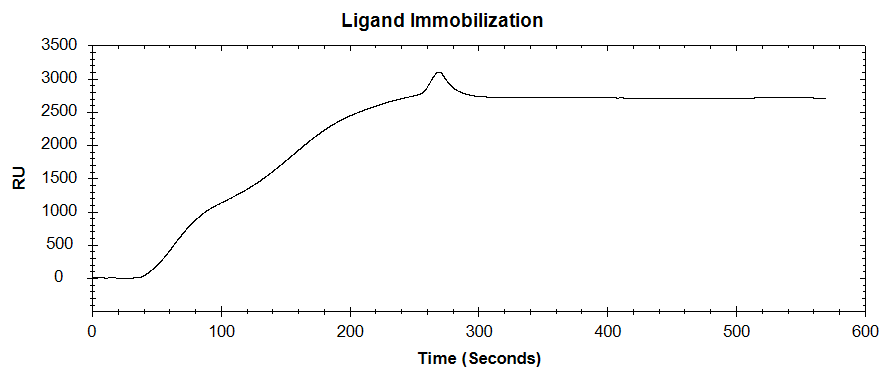
In our latest application note, kinetic binding of a SARS-CoV-2 monoclonal antibody (mAb) to the SARS-CoV-2 spike protein receptor-binding domain (RBD) was measured using the OpenSPR-XT™. This was done with both the primary mAb in buffer, and the same mAb in diluted serum. Using a secondary antibody amplification technique, a picomolar limit of detection (LOD) for the primary antibody was obtained. The RBD was immobilized to a carboxyl sensor by following a standard amine coupling procedure (Figure 1). Bovine Serum Albumin (BSA) was used to prevent non-specific binding. The RBD was immobilized to a carboxyl sensor by following a standard amine coupling procedure (Figure 1). Bovine Serum Albumin (BSA) was used to prevent non-specific binding.
In one experiment (Figure 2), primary mAb was diluted in the running buffer while in another experiment (Figure 3) the same dilutions were performed in 50% serum, 50% running buffer. In both cases, the concentration levels were the same and ran in ascending order, followed by a fixed concentration of the amplification (secondary) mAb, followed by a regeneration step. Both data sets were analyzed in TraceDrawer using a 1:1 fitting model and the resulting affinity and kinetics are shown in Table 1.
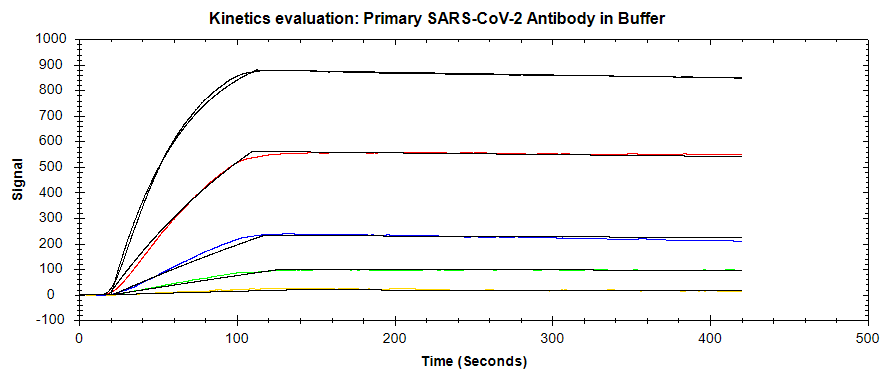
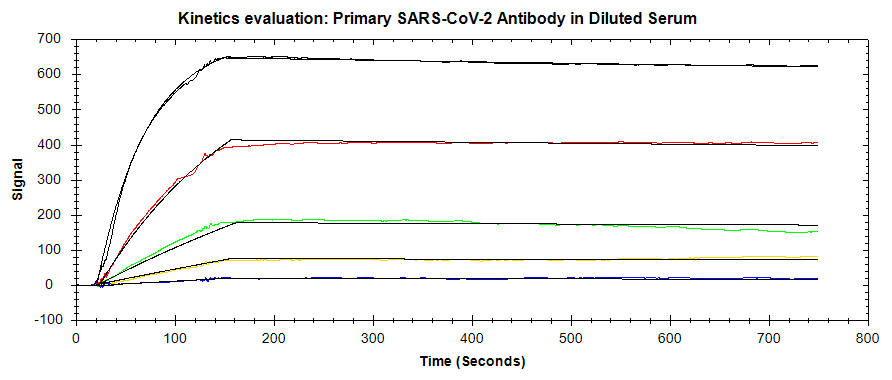
Table 1: Kinetic and affinity values determined using OpenSPR™ data with the TraceDrawer™ analysis software.
| Parameter | SARS-CoV-2 mAb in buffer | SARS-CoV-2 mAb in Serum |
| ka [1/M*s] | 1.51e5 (±2.17e0) | 1.30e5 (±3.94e0) |
| kd [1/s] | 1.20e-4 (±1.60e-5) | 6.75e-5 (±2.00e-5) |
| KD | 7.93e-10 (±1.06e-10) | 5.18e-10 (±1.53e-10) |
OpenSPR-XT™ for Measuring Binding Affinity and Kinetics of SARS-CoV-2 Antibody
Using the OpenSPR-XT™, the binding of SARS-CoV-2 monoclonal antibody to the SARS-CoV-2 spike protein receptor-binding domain was successfully characterized in a high-serum background, with comparable results to a standard kinetic assay without any serum. This demonstrates the specificity and high affinity of this antibody, indicating its utility in a variety of potential applications. It also illustrates the efficacy of OpenSPR to be used in screening applications where low quantities of antibody are present in a complex medium. The OpenSPR™ is able to quantify a diverse range of biomolecular interactions in a variety of conditions, making it an excellent tool for studying COVID-19, and other viruses.
While many researchers face challenges in their research programs due to COVID-19 related closures, continuing to have access to crucial instrumentation that allows for the study of viruses and other disease agents is essential. We have a robust and well established remote process to provide product demonstration, support, installation, and training for users, so they can succeed in spite of the travel restrictions and closures. The OpenSPR™ system gives any researcher the ability to study quantitative binding kinetics right from their own lab bench.
Supporting Researchers During COVID-19
We’re here to support your research during COVID-19. Regardless of if you are a Nicoya customer yet or not – if you would benefit from an SPR expert’s feedback on a data set or are currently designing an SPR experiment, our Customer Success Scientists are happy to provide an evaluation. We can then arrange a follow-up call to discuss our findings. Together, we can keep research moving forward and prioritize improving human life. Get an SPR expert’s opinion here.
