Surface plasmon resonance (SPR) can be used to analyze all types of interactions ranging from protein-protein, protein-small molecule, protein-nucleic acid, protein-aptamers, protein-lipids, and many more. One area of research is the study of lipid binding kinetics, and many scientists are implementing techniques such as SPR to define the affinity and specificity of these interactions.
Why do we study lipid interactions?
Can you guess how many different lipids cell membranes are made up of? More than 1000! These lipids direct signalling processes in cells through target proteins, which means that lipid-protein interactions are important for signalling and trafficking processes in cells. Nearly half of all proteins are located in or on the membrane, so there are plenty of lipid-protein interactions that need to be characterized!
Why is determining lipid specificity and affinity important?
High-affinity interactions between peripheral proteins regulate lipid signalling from the cellular membrane. Using techniques like surface plasmon resonance (SPR) to accurately determine lipid specificity and membrane affinity is important for establishing how the membrane interface signals throughout a cell. There are a few biochemical and biophysical experiments used to determine the specificity and affinity of lipids in the field, but SPR has become a key label-free technique for this type of analysis.
Which lipid sensor chip do I use to immobilize my lipid to the sensor?
SPR is a label-free technology which allows researchers to quantitatively analyze binding between two biomolecules. SPR technology allows us to determine the kon, koff and KD of interactions, providing deeper insight into binding events compared to other techniques that only give endpoint measurements, such as pull-down assays. There are two sensor chips available at Nicoya for immobilizing lipid ligands to the sensor surface.
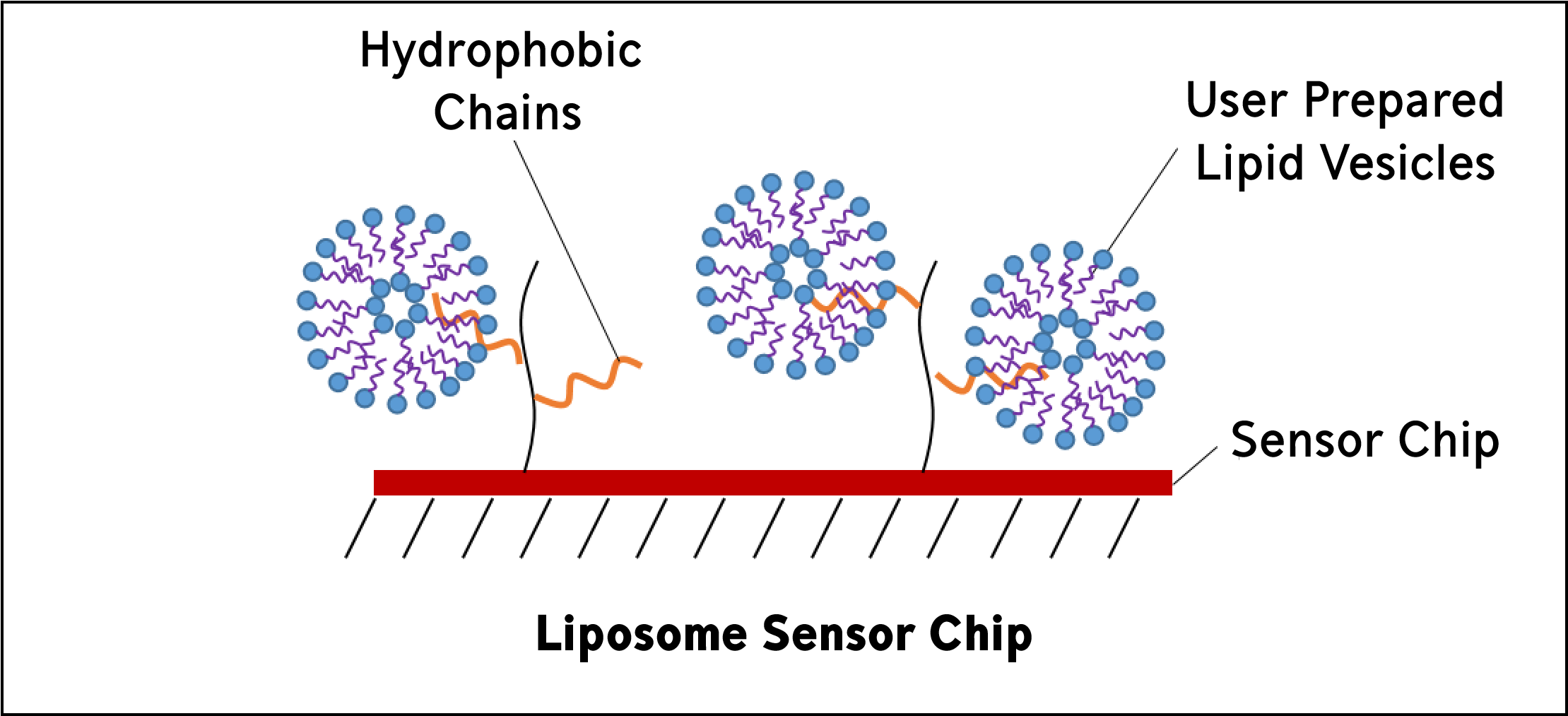
Liposome sensor chips are used to capture user-prepared liposomes. They are composed of covalently attached lipophilic groups. Lipid vesicles, or liposomes, mimic biological membranes and can be a convenient way to study membrane systems.
Yes, this means that you can study transmembrane proteins using liposome sensor chips by reconstituting your protein into the liposome and directly capturing the liposome using the liposome sensor chips!
Uses:
Liposome-protein interactions
Membrane protein-protein interactions
Peripheral protein-protein interactions
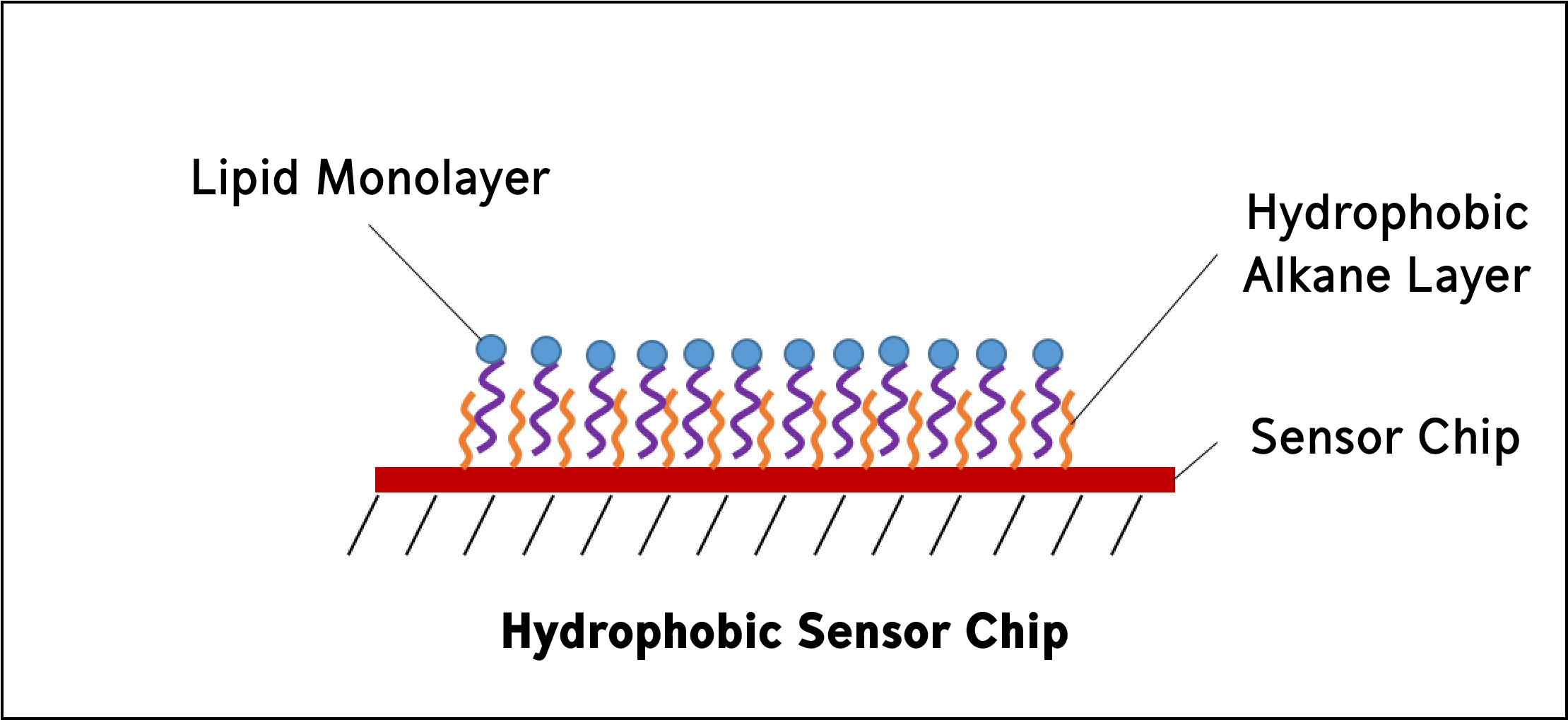
Hydrophobic sensor chips are used to immobilize lipid molecules into a monolayer on the sensor surface. Hydrophobic sensor chips are coated with a hydrophobic layer of long-chain alkane molecules, which means that partially solubilized lipids will self-assemble to the surface creating a monolayer with the hydrophilic groups positioned away from the surface.
Uses:
Lipid-protein interactions
Lipid-lipid interactions
Example of a lipid-protein interaction analyzed using OpenSPR
Below is an example of how OpenSPR™was used to characterize the binding kinetics of a protein binding to two different lipids. This study identifies how the location of the phosphate group location on the lipids impacts the binding kinetics. This data illustrates the value of surface plasmon resonance analysis in such applications. You can find the full study here.
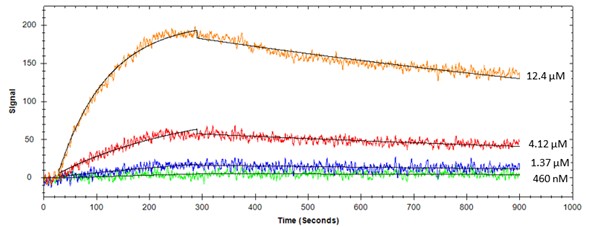
Figure 1. Protein binding to immobilized liposome #1 on OpenSPR
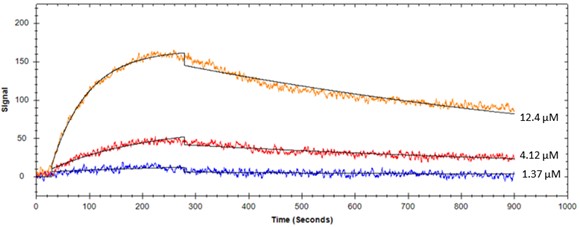
Figure 2. Protein binding to immobilized liposome #2 on OpenSPR
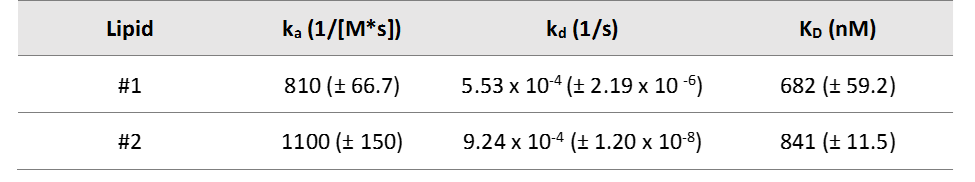
Table 1. Kinetic constants determined using SPR for protein-lipid interactions
V. Hodnik and G. Anderluh., “Surface plasmon resonance for measuring interactions of proteins with lipid membranes.” Methods Mol Biol, Vol. 974, pp. 23-36, 2013.
