Overview
The analysis of binding kinetics with SPR plays a critical role in the understanding of COVID-19. This includes basic research for understanding the viral infection mechanisms, the development of therapeutics & vaccines, and the realization of effective diagnostic tools. Although RT-PCR is the primary method of COVID-19 testing currently used, it requires dedicated labs and trained operators, and the supply chain is struggling to keep up with test demand. Direct viral detection through the use of high-affinity antibodies could offer an alternative screening approach due to its speed and simplicity. Also, serological testing for COVID-19 specific antibodies can be used as a diagnostic tool for those that no longer have the virus, and could be used to indicate immunity. The presence of viral-specific antibodies in the host can be used as a diagnostic tool to aid the medical practitioners in determining the effectiveness of vaccines as well as identify high-risk targets, such as those with under-responsive immune systems that may need more intensive treatments.
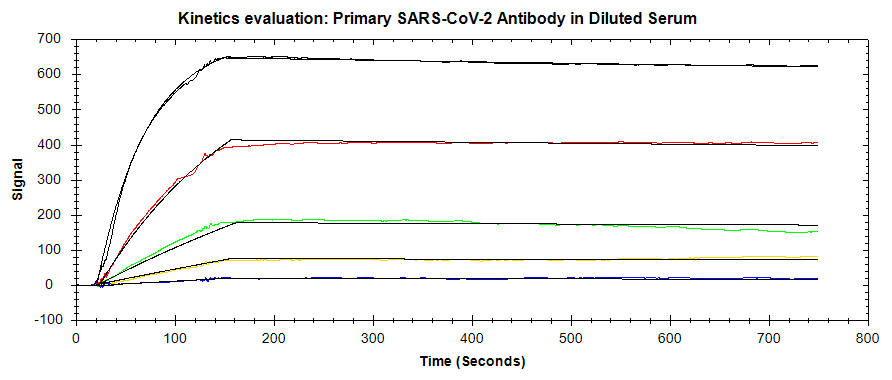
In this application note, the OpenSPR-XT™ was used to detect and measure the binding kinetics of the SARS-CoV-2 monoclonal antibody to the SARS-CoV-2 spike protein RBD. A direct binding assay was performed to measure the kinetics of this interaction, in a normal buffer background as well as by spiking the antibody into 50% serum. The LOD for the primary antibody was also optimized through secondary antibody amplification. The binding signal obtained indicates that the presence of the SARS-CoV-2 antibody could be detected down to picomolar concentrations. The OpenSPR-XT™ is an affordable and user-friendly benchtop SPR instrument that enables the measurement of label-free binding kinetics of diverse biomolecular interactions.
